Isocitrate dehydrogenase mutation as a therapeutic target in gliomas
Introduction
There have been major advances in the understanding of isocitrate dehydrogenase (IDH) mutations in various types of cancers since their discovery in 2008 by Parsons et al. (1). In their integrated genomic analysis of human glioblastoma (GBM), IDH mutation was observed in 12% of GBM samples and the majority of these were in tumors that had progressed from lower grade gliomas (secondary GBM). Subsequently >70% of WHO grade II/III gliomas were found to harbor an IDH mutation (2). IDH mutations also occur in acute myeloid leukemia (AML), myelodysplastic syndrome (MSD), myeloproliferative neoplasm (MPN), cholangiocarcinoma, enchondroma and chondrosarcoma, and other solid cancers. Significant efforts have been made since its discovery to improve our knowledge about the role IDH mutations play in tumorigenesis and progression. It has become clear that IDH mutations are associated with many epigenetic and metabolic changes in these tumors. In this review we provide an overview of the function of mutant IDH and the current understanding of the role IDH mutations play in tumorigenesis. In addition, we discuss several potential therapeutic strategies for IDH-mutant gliomas based on emerging preclinical and clinical data, including therapies targeting mutant IDH and other IDH mutation-associated pathways.
Functions of normal IDH
There are three isozymes in the IDH family: IDH1 is located in the cytosol and peroxisome and IDH 2 and IDH3 are located in the mitochondria. Normal IDH catalyzes the NADP+-dependent oxidative decarboxylation of isocitrate to α-ketoglutarate (α-KG), producing CO2 and NADPH in the process (3). It, therefore, plays an important role in biosynthesis of central metabolites in the tricarboxylic acid (TCA) cycle as well as in the generation of cellular NADPH. IDH1 is particularly important in the brain as this is the main source of NADPH (4).
Functions of mutant IDH
The majority of IDH mutations involve the catalytic pocket of the enzyme. IDH1 mutations predominantly occur at arginine 132 resulting in substitutions, including R132H (most common, 88%), R132C, R132L, R132S, and R132G (2). IDH2 mutations typically occur at R140 or R172. Of IDH2 mutations, R172K is most common. IDH1 and IDH2 mutations are mutually exclusive.
A landmark study of a large-scale metabolomics screen in IDH-mutant GBM cells found that IDH1R132H mutation leads to a neomorphic ability to produce (R)-2-hydroxyglutarate (2HG) while oxidizing NADPH to NADP+ (5). This process results in 2HG accumulation with levels up to 100-fold higher in IDH mutant versus IDH wild type cells (6). 2HG is converted directly from α-KG by the mutant IDH, therefore relies on the presence of a wild-type IDH which converts isocitrate to α-KG in the TCA cycle (7). This may explain the rareness of loss of heterozygosity of IDH.
2HG is considered as an ‘oncometabolite’ which is thought to be responsible for many, if not all, biologic effects of IDH mutations. In normal cells without IDH mutation, 2HG is present at trace levels and has no known metabolic function (8). It is rapidly converted back to α-KG by 2HG dehydrogenases. In IDH-mutant cells, the production of 2HG is far greater than its clearance, leading to high concentrations of 2HG in these cells.
Role of IDH mutations in tumor formation and progression
IDH mutations have been suggested as an important early event in tumorigenesis. Large scale genomic analyses of IDH-mutant gliomas found that IDH1 mutation occurred before acquisition of other mutations such as TP53 mutations or 1p/19q co-deletion (9,10), and IDH1R132H was the only mutation that was consistently present in both the initial and recurrent glioma samples (11). IDH mutation was also maintained in 95% of recurrent secondary GBMs (12). Further supporting evidence of IDH mutation being an early genetic event comes from studies of hematopoietic disorders where IDH1/2 mutations have been found in premalignant diseases such as early MDS, MDS/MPN (13-15). IDH mutations have also been identified in other premalignant solid tumors, such as enchondromas, cartilaginous tumors that can progress to malignant chondrosarcomas.
It remains unclear whether IDH mutation by itself causes cancer or if it requires other oncogenic events to initiate tumorigenesis. IDH-mutant tumors typically harbor other cancer-associated genetic alterations. IDH mutation is associated with ATRX, TP53 and TERT mutations in diffuse astrocytoma, and co-deletion of chromosome arms 1p and 19q in oligodendroglioma (16). In mouse IDH-mutant AML models, there was a long latency and incomplete penetrance of tumors, suggesting IDH mutation is not sufficient to fully drive tumorigenesis by itself (17-19). Similarly, IDH1R132H expression in several cell types of the murine central nervous system failed to form tumor on its own (20). However, once established, at least in IDH1-mutant glioma xenograft models in mice, ongoing expression of IDH1 mutation appears to be necessary for tumor maintenance as pharmacological inhibition of mutant IDH1 results in tumor growth delay and induction of cell differentiation (21).
Epigenetic modifications by IDH mutations via 2HG accumulation
2HG is structurally similar to α-KG. At high concentrations, 2HG competitively inhibits α-KG-dependent dioxygenases including the Jumonji domain containing (JmjC) family of histone lysine demethylases (KDMs) and the ten-eleven translocation (TET) family of DNA hydroxylases that catalyze the sequential oxidation of 5-methylcytosine (5mC) to eventual DNA demethylation (22-27). These inhibitory effects on histone and DNA demethylases result in a hypermethylation phenotype with alterations in cellular epigenetic status. IDH-mutant tumors have been shown to display a CpG island methylator phenotype (CIMP) (28,29). In glioma, this DNA hypermethylation signature is known as glioma CIMP (G-CIMP). The notion that IDH mutation is the molecular basis of G-CIMP has been supported by preclinical data showing that introduction of mutant IDH1 into primary human astrocytes induces extensive DNA hypermethylation and reshapes the methylome in a fashion that mirrors the changes observed in G-CIMP (30).
IDH mutations and subsequent accumulation of 2HG have been shown to prevent the histone demethylation that is required for lineage-specific progenitor cells to differentiate (31). This suggests that IDH mutations can promote tumorigenesis by blocking cellular differentiation via hypermethylation of genes involved in differentiation.
CTCF insulator protein organizes the genome into topological domains by creating chromatin loops and boundaries (32,33). The hypermethylator phenotype of IDH-mutant gliomas has been shown to disrupt CTCF binding and domain boundaries, allowing a potent constitutive enhancer to interact with the normally quiescent PDGFRA oncogene promoter (34). This provides an epigenetic mechanistic link between IDH mutation and activation of PDGFRA. Furthermore, a demethylating agent was able to restore the insulator function, resulting in reduction of PDGRFA expression in IDH-mutant astrocytoma model. Widespread CTCF and domain boundary disruption is likely and this could lead to other oncogene activations in IDH-mutant tumors.
Potential therapeutic targets in IDH-mutant gliomas
Patients with WHO grade II and III gliomas that harbor an IDH mutation have a better response to treatment and a prolonged survival compared to those with IDH wild-type gliomas (35,36). In addition, these lower grade gliomas respond better to chemo-radiotherapy than to radiotherapy alone (37,38). IDH mutant gliomas maintain the mutant IDH allele even after acquiring further oncogenic driver mutations (11,39). This may indicate that IDH mutant gliomas, even at progression or after malignant transformation to higher grade glioma, may remain vulnerable to the targeted therapies developed specifically for IDH mutations. The therapeutic effects may be further enhanced by combining different targeted therapies or with traditional chemotherapeutics or radiation. In this section, various potential therapeutic targets in IDH-mutant gliomas are discussed based on emerging preclinical and clinical data.
Mutant IDH enzymes
The notion that these neomorphic mutations of IDH1 and IDH2 are the mechanistic basis for malignant transformation led to the development of drugs targeting the mutant IDH1/2 enzymes. Moreover, IDH mutations are tumor-specific and expressed uniformly in all tumor cells (10). In a preclinical study using a fully transformed cells with endogenous IDH1 mutation, a targeted inhibitor of mutant IDH1 induced demethylation of histone H3K9me3 and expression of genes associated with gliogenic differentiation via reduction of 2HG production (21). Mutant IDH1 inhibition also delayed the growth of IDH1-mutant, but not IDH1 wildtype, glioma cells. In another study, a mutant IDH2 inhibitor was able to induce cellular differentiation of primary human AML cells harboring an IDH2 mutation (40). Mechanistic studies in an IDH2 mutant erythroleukemia model showed that inhibition of IDH2 by a small molecule inhibitor reversed both histone and DNA methylation changes within days and weeks, respectively (41). Methylation of histone and DNA is closely linked to mRNA expression and differentiation, therefore these preclinical study results provide proof-of-concept that targeting mutant IDH1/2 is a potential therapeutic strategy for IDH-mutant tumors.
Whether inhibition of mutant IDH and subsequent reduction in 2HG production are sufficient to halt tumor growth in IDH-mutant gliomas and other solid tumors remains unclear. There are studies that have shown no or minimal effects on CIMP or tumor growth despite near complete abrogation of 2HG production by pharmacologic inhibition alone (42-45). These conflicting observations between tumor types and cell lines may be due to potentially differing effector pathways of mutant IDH and 2HG in cellular-, (epi)genetic-, and organ-level context.
Multiple IDH inhibitors have been developed and several of them are currently undergoing clinical evaluation in phase I/II trials in patients with IDH mutant tumors. AG-221, a selective small molecule inhibitor of IDH2 was the first to enter clinical trial in late 2013. Preliminary data from phase I trial of this agent in patients with IDH2-mutant relapsed or refractory AML demonstrated clinical responses in about 40% of the patients (46). Preliminary data of the dose escalation phase I trial of AG-120, an IDH1 inhibitor, in solid tumors reported a clinical benefit rate (defined as progression free at 6 months) of 37% among all patients and 25% in glioma patients (47). AG-881, a pan-inhibitor of the mutant IDH1 and IDH 2 enzymes, has been shown to fully penetrate the brain-blood barrier in preclinical studies, therefore it is of particular interest whether this agent has better clinical efficacy in glioma patients. At the time of writing, there are six IDH inhibitor clinical trials for gliomas (Table 1).
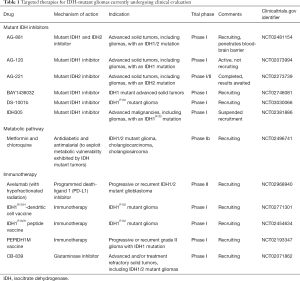
Full table
DNA hypermethylation
Given the evidence that IDH mutations cause epigenetic alterations and a cellular differentiation block by inhibiting histone and DNA demethylases and the implication that these epigenetic alterations lead to malignant transformation, histone deacetylase inhibitors and DNA demethylating agents have been investigated in preclinical models of IDH-mutant tumors (42,45,48). Decitabine and azacytidine are DNA demethylating agents, approved by Food and Drug Administration (FDA) for MSD, and they inhibit DNA (cytosine-5-)-methyltransferase 1 (DNMT1). A preclinical study of decitabine in patient-derived IDH1 mutant glioma cells demonstrated that decitabine can effectively reverse the pathologic DNA hypermethylation induced by mutant IDH, and induce cellular differentiation in these cells (45). In this study, decitabine also decreased growth of these cells in vitro and IDH1-mutant xenografts in mice. Similarly, another study showed that long-term administration of low dose azacytidine results in reduction of DNA methylation of promoter loci, induction of glial differentiation, reduction of cell proliferation and a significant reduction in tumor growth in an endogenous IDH1 anaplastic astrocytoma model (42). The effects of both decitabine and azacytidine were durable even after the treatment was stopped.
These demethylating agents may be useful not only in the recurrent setting but also as a maintenance therapy following surgical resection of IDH-mutant gliomas. Due to the infiltrative growth pattern of gliomas it is almost impossible to remove all of the tumor cells by resection, therefore tumor recurrence is virtually inevitable over time despite gross total resection and adjuvant chemo-radiotherapy, and often these tumors recur as higher grade gliomas. Long-term low dose treatment with a DNA demethylating agent following initial treatment may induce cell differentiation in the remaining IDH-mutant glioma cells, thereby delaying or even preventing recurrence. Targeting the pathologic DNA hypermethylation in IDH-mutant cells with DNA demethylating agents may represent a potential therapeutic strategy that warrant further clinical evaluation in patients with IDH-mutant gliomas.
Vulnerability of IDH-mutant cells to NAD+ depletion
IDH mutations have also been shown to affect canonical metabolic pathways including NAD+ production, glutamine catabolism and the TCA cycle (3,43,44,49-51). Tateishi et al. reported intriguing data showing that depletion of 2HG via direct inhibition of mutant IDH1 was insufficient to halt the formation or growth of progressive IDH1-mutant gliomas in vitro and in vivo in mice (44). This finding suggests that other genetic and/or epigenetic factors may modulate the effect of direct mutant IDH1 inhibition. The authors then discovered that IDH-mutant cells were more sensitive, compared to IDH wild-type cells, to depletion of the coenzyme NAD+, an essential component of intracellular signaling pathways implicated in cancer cell growth (52). Depletion of NAD+ using small molecule inhibitors of NAMPT, a salvage NAD+ synthesis enzyme, resulted in selective cytotoxicity in IDH-mutant glioma cells, and in mice harboring intracerebral IDH-mutant glioma xenografts, treatment with NAMPT inhibitors resulted in a marked survival extension (44). They also found that IDH1 mutation led to reduced expression of Naprt1, a rate-limiting enzyme in the salvage NAD+ pathway, via 2HG-mediated hypermethylation of the NAPRT1 promoter CpG island. This explains the lower basal levels of NAD+ found in IDH-mutant cells which may render them less able to compensate for further depletion of NAD+ induced by NAMPT inhibition. A metabolic study of IDH-mutant glioma cell lines treated with NAMPT inhibitors showed that NAD+ depletion disrupts TCA cycle metabolism and induces autophagy by activating AMP-activated protein kinase (AMPK), a key regulator of the metabolic checkpoint which responds to nutrient- and energy-poor conditions (44). Collectively, these data suggest that NAMPT inhibition represents a promising synthetic lethal therapeutic approach that effectively inhibits both NAD+ salvage pathways (NAMPT and Naprt1), leading to a metabolic crisis that activates AMPK and initiates autophagy in IDH-mutant gliomas.
IDH mutation-associated genome instability
Mutant IDH uses NADPH as a cofactor in the conversion of α-KG to 2HG, resulting in alterations in the NADPH/NADP+ ratio. A significantly reduced NADPH production has been reported in IDH-mutant glioma samples (4). NADPH is an essential reducing factor required for cellular defense mechanism against reactive oxygen species through regeneration of reduced glutathione (GSH) from oxidized glutathione (GSSG) by glutathione reductase. In IDH-mutant cell lines, IDH mutation resulted in increased DNA double-strand breaks after ionizing radiation via reduction of IDH-mediated NADPH production (53). This observation suggests that IDH mutations lead to DNA damage and genome instability, which in turn render these IDH-mutant cells more sensitive to radiation. Mutant IDH inhibitor restored IDH-mediated NADPH production with a subsequent radioresistance of IDH-mutant cells, suggesting that treatment with mutant IDH inhibitor during radiation therapy may abolish the beneficial effects on survival of IDH-mutant glioma patients.
Alkylating agents, such as temozolomide, procarbazine and lomustine, are commonly used chemotherapeutics in glioma patients. Chemotherapy-induced DNA lesions are repaired by DNA repair enzymes such as O6-methylguanine-DNA methyltransferase (MGMT) and alkB homolog (ALKBH) proteins (54-56). The G-CIMP status correlates well with MGMT promoter methylation in gliomas (30), suggesting a link between IDH mutation and MGMT methylation. ALKBHs are α-KG dependent, therefore, 2HG accumulation in IDH-mutant cells inhibit the function of these DNA repair proteins, leading to increased DNA double-strand breaks after alkylating treatment (56). As a result, IDH mutations sensitize IDH-mutant tumor cells to alkylating agents compared to IDH wildtype.
Recently IDH1/2 mutations have been shown to cause a homologous recombination (HR) defect via 2HG-induced inhibition of α-KG-dependent dioxygenases, particularly KDM4A and KDM4B (57). This “BRCAness” phenotype was demonstrated in a range of clinically relevant models, including patient-derived IDH-mutant glioma cell lines, and these cells showed a selective sensitivity to poly(adenosine 5'-diphosphate-ribose) polymerase (PARP) inhibition via synthetic lethal interactions. Treatment with a mutant IDH inhibitor reversed this HR defect and eliminated the associated PARP inhibitor sensitivity. Olaparib and veliparib, PARP inhibitors, have been shown to penetrate the blood-brain barrier (58-60). PARP inhibition using these drugs as a potential treatment for IDH-mutant gliomas warrant further investigation.
Collectively, these studies provide evidence that IDH mutations are associated with increased genome instability, thereby greater vulnerability to therapies causing further DNA damage or interfering with DNA repair. This may explain, at least partly, the more favorable therapeutic responses and better prognosis observed in IDH-mutant gliomas (37,61).
Dependence of IDH-mutant cells on oxidative mitochondrial metabolism
IDH1 mutations have been shown to compromise cellular metabolism under hypoxic conditions in IDH1-mutant cell lines, resulting in increased dependence on oxidative mitochondrial metabolism and inability to induce reductive glutamine metabolism (43). This suggests that IDH-mutant cells may be more sensitive to inhibitors that perturb mitochondrial metabolism. Interestingly, inhibition of mutant IDH1 was unable to reverse the metabolic phenotype, a defect in reductive carboxylation activity, suggesting that the metabolic alterations may be independent of 2HG production (43).
Further evidence that IDH-mutant cells are vulnerable to mitochondrial metabolism inhibition was provided by a preclinical metabolomics study of IDH1R132H-mutant cells (62). Metabolic reprogramming of IDH1-mutant cells resulted in an inability of these cells to generate energy from various carbon substrate fuels. Treatment with metformin, a mitochondrial biguanide poison, decreased cell growth and survival of IDH1-mutant cells by altering the oxidative metabolism of glutamine and glucose via the TCA cycle. In addition, this study also showed that the metabolic vulnerability of IDH1-mutant cells to metformin appeared independent of 2HG production.
The importance of mitochondria on the growth of IDH-mutant tumors was also suggested by another study in which IDH1 mutant glioma samples and IDH1R132H-expressing cancer cells were found to be hypersuccinylated, preferentially in the mitochondria (63). 2HG was shown to inhibit the mitochondrial TCA cycle enzyme succinate dehydrogenase (SDH) resulting in mitochondrial respiration impairment via succinyl-CoA accumulation and hypersuccinylation. This mitochondrial dysfunction induced anti-apoptotic protein BCL-2 accumulation at the mitochondria membrane, leading to apoptosis resistance in these cells. Overexpression of a desuccinylase SIRT5 was shown to reverse these processes and inhibit growth of hypersuccinylated tumors.
There is currently one phase I trial exploring the combination of metformin and chloroquine in IDH-mutant tumors including gliomas (Table 1). Further investigations exploring the combination of mutant IDH inhibition and oxidative mitochondrial metabolism inhibition as a potential therapeutic strategy for IDH-mutant tumors are warranted.
α-KG homeostasis via glutaminase inhibition
2HG has been shown to primarily drive from glutamine, where glutamine is hydrolyzed by glutaminase to produce glutamate which is converted to α-KG (5). α-KG is then converted to 2HG by mutant IDH. A small molecule glutaminase inhibitor, BPTES, has been shown to reduce glutaminase activity with subsequent reduction in glutamate and α-KG levels in both IDH-mutant and IDH-wildtype GBM cells, but interestingly, only the growth of IDH mutant cells was slowed (64). The growth reduction seen in this study was modest (about 20% reduction) although the reduction of glutaminase activity was significant, indicating that cellular metabolism is able to compensate for changes made by glutaminase inhibition. Glutaminase inhibition alone, therefore, is unlikely to be an effective therapy, however, it may exert synergistic effects on tumor growth when combined with other therapeutic strategies. Another potent glutaminase inhibitor, CB-839, is currently undergoing phase I evaluation in various solid tumors including IDH mutant tumors (Table 1).
IDH mutation-associated hypoxia-inducible factor-1α (HIF-1α) pathway
α-KG is essential for prolyl hydroxylases (PHD) which promotes HIF-1α degradation therefore a reduction in α-KG, as in IDH mutant cells, abrogate degradation of HIF-1α. In glioma cell lines, IDH1 knockdown induced the expression of HIF-1α target genes, including glucose transporter 1 (Glut1), vascular endothelial growth factor (VEGF), and phosphoglycerate kinase 1 (PGK1), that are implicated in glucose metabolism, angiogenesis, and other signaling pathways that are critical to tumor growth (65). Consistent with this, IDH1 mutations correlated with elevated HIF-1α and VEGF levels in human glioma samples. Similarly, another preclinical study also found increased HIF-1α and cell proliferation in IDH-mutant glioma cell lines (66). In addition, this study showed that IDH mutation activated nuclear factor-κB (NF-κB) in a HIF-1α dependent manner, which induced high expression levels of oncogenes, cyclins D1 and E and c-Myc in these cells. These results suggest that IDH mutations contribute to tumor angiogenesis and proliferation by promoting the HIF-1α pathway. In cultured cells expressing IDH1 mutation, it has been shown that an α-KG derivative reversed the induction of HIF-1α levels (65). Drugs targeting HIF-1α or drugs mimicking α-KG may thus warrant further investigation as a potential therapy for IDH-mutant gliomas.
Mammalian target of rapamycin (mTOR) signaling pathway
KDM4 is a subfamily of the Jumanji family of histone demethylases and is inhibited by 2HG via α-KG inhibition (25). 2HG-induced KDM4 inhibition was recently shown to promote oncogenic activation of mTOR independently of the PI3K/AKT/TSC1–2 pathway by reducing the levels of DEPTOR (67), a negative regulator of both mTORC1 and mTORC2 which are associated with cell growth and autophagy (mTORC1), and cell survival and cytoskeleton organization (mTORC2) (68). This mTOR activation via the mutant IDH/2HG/KDM4/DEPTOR pathway may provide an additional molecular mechanism to explain the oncogenic activity of IDH1/2 mutant gliomas and represents a potential therapeutic strategy.
Immunotherapy
Given the evidence that IDH mutation is an early event in tumorigenesis and is present homogenously in all glioma tumor cells at specific codons, these mutations are ideal immunotherapy targets, especially as a maintenance therapy for preventing recurrence of these diffuse infiltrating tumors. Schumacher et al. demonstrated that IDH1R132H contained an immunogenic epitope suitable for mutation-specific vaccination (69). Immunization with mutated IDH1 peptides resulted in a significant prolongation of survival in an intracranial glioma mouse model (70). Immunotherapy using peptide and dendritic cell vaccinations are currently being investigated in patients with IDH-mutant gliomas (Table 1).
Natural killer (NK) cells are critical to the innate immune system and respond to tumor formation. Lysis of tumor cells by NK cells is mediated by interaction between NK group 2D (NKG2D) receptor on NK cells and stress-inducible ligands on target cells, such as NKG2D ligand (NKG2D-L) (71). Correlation between NKG2D-L expression and improved clinical outcomes have been shown in human cancers (72). Zhang et al. discovered that IDH-mutant glioma cells were resistant to NK-mediated lysis due to transcriptional silencing of NKG2D-L by 2HG-induced genomic hypermethylation (71). Decitabine upregulated the expression of NKG2D-L and sensitized the cells to NK cell-mediated cytotoxicity.
In the IDH1-vaccine study by Schumacher et al., only 4 of 25 patients with IDH-mutant gliomas exhibited evidence of spontaneous T-cell response to the IDH1 epitope (69). This may be explained by the impaired innate immune activity shown by Zhang et al., as innate immune activity is required for induction and maintenance of T-cell mediated immunity. Demethylating agents may therefore facilitate potent antitumor T-cell mediated immune responses of vaccine therapy for IDH-mutant gliomas. Further investigations in clinical trials are warranted for this combined approach.
Conclusions
An IDH mutation is found in the vast majority of low grade gliomas and secondary GBM and it appears to be an early event. Significant efforts have been made to better understand the role IDH mutations play in tumor initiation, maintenance and progression. A wide range of epigenetic and metabolic alterations caused by mutant IDH-mediated 2HG accumulation via inhibition of α-KG-dependent dioxygenases have been implicated, and many different potential therapeutic avenues are being explored. Specific aberrant changes, that drive tumor formation and growth, may differ in different tumor types, further complicating identification of therapeutic targets in these tumors. Drug combinations that exert synergistic effects on more than one target simultaneously may be more effective. Finally, although a matter of controversy IDH mutations represent a “hit and run” genetic event in gliomas potentially explaining the general lack of efficacy of first-generation IDH1 inhibitors and suggesting that other synthetic lethal or immune-targeting strategies may be more effective.
Acknowledgements
This article was supported by the National Cancer Institute of the National Institutes of Health under Award Number P50CA165962 (to TT Batchelor).
Footnote
Conflicts of Interest: TT Batchelor reports consulting relationships with Amgen, NXDC, Merck, and UpToDate. The other author has no conflicts of interest to declare.
References
- Parsons DW, Jones S, Zhang X, et al. An integrated genomic analysis of human glioblastoma multiforme. Science 2008;321:1807-12. [Crossref] [PubMed]
- Yan H, Parsons DW, Jin G, et al. IDH1 and IDH2 mutations in gliomas. N Engl J Med 2009;360:765-73. [Crossref] [PubMed]
- Leonardi R, Subramanian C, Jackowski S, et al. Cancer-associated isocitrate dehydrogenase mutations inactivate NADPH-dependent reductive carboxylation. J Biol Chem 2012;287:14615-20. [Crossref] [PubMed]
- Bleeker FE, Atai NA, Lamba S, et al. The prognostic IDH1(R132) mutation is associated with reduced NADP+-dependent IDH activity in glioblastoma. Acta Neuropathol 2010;119:487-94. [Crossref] [PubMed]
- Dang L, White DW, Gross S, et al. Cancer-associated IDH1 mutations produce 2-hydroxyglutarate. Nature 2009;462:739-44. [Crossref] [PubMed]
- Gross S, Cairns RA, Minden MD, et al. Cancer-associated metabolite 2-hydroxyglutarate accumulates in acute myelogenous leukemia with isocitrate dehydrogenase 1 and 2 mutations. J Exp Med 2010;207:339-44. [Crossref] [PubMed]
- Jin G, Reitman ZJ, Duncan CG, et al. Disruption of wild-type IDH1 suppresses D-2-hydroxyglutarate production in IDH1-mutated gliomas. Cancer Res 2013;73:496-501. [Crossref] [PubMed]
- Dang L, Yen K, Attar EC. IDH mutations in cancer and progress toward development of targeted therapeutics. Ann Oncol 2016;27:599-608. [Crossref] [PubMed]
- Lai A, Kharbanda S, Pope WB, et al. Evidence for sequenced molecular evolution of IDH1 mutant glioblastoma from a distinct cell of origin. J Clin Oncol 2011;29:4482-90. [Crossref] [PubMed]
- Watanabe T, Nobusawa S, Kleihues P, et al. IDH1 mutations are early events in the development of astrocytomas and oligodendrogliomas. Am J Pathol 2009;174:1149-53. [Crossref] [PubMed]
- Johnson BE, Mazor T, Hong C, et al. Mutational analysis reveals the origin and therapy-driven evolution of recurrent glioma. Science 2014;343:189-93. [Crossref] [PubMed]
- Lass U, Numann A, von Eckardstein K, et al. Clonal analysis in recurrent astrocytic, oligoastrocytic and oligodendroglial tumors implicates IDH1- mutation as common tumor initiating event. PLoS One 2012;7:e41298. [Crossref] [PubMed]
- Kosmider O, Gelsi-Boyer V, Slama L, et al. Mutations of IDH1 and IDH2 genes in early and accelerated phases of myelodysplastic syndromes and MDS/myeloproliferative neoplasms. Leukemia 2010;24:1094-6. [Crossref] [PubMed]
- Pardanani A, Lasho TL, Finke CM, et al. IDH1 and IDH2 mutation analysis in chronic- and blast-phase myeloproliferative neoplasms. Leukemia 2010;24:1146-51. [Crossref] [PubMed]
- Thol F, Weissinger EM, Krauter J, et al. IDH1 mutations in patients with myelodysplastic syndromes are associated with an unfavorable prognosis. Haematologica 2010;95:1668-74. [Crossref] [PubMed]
- Network TCGAR. Comprehensive, Integrative Genomic Analysis of Diffuse Lower-Grade Gliomas. N Engl J Med 2015;372:2481-98. [Crossref] [PubMed]
- Chen C, Liu Y, Lu C, et al. Cancer-associated IDH2 mutants drive an acute myeloid leukemia that is susceptible to Brd4 inhibition. Genes Dev 2013;27:1974-85. [Crossref] [PubMed]
- Kats LM, Reschke M, Taulli R, et al. Proto-oncogenic role of mutant IDH2 in leukemia initiation and maintenance. Cell Stem Cell 2014;14:329-41. [Crossref] [PubMed]
- Sasaki M, Knobbe CB, Munger JC, et al. IDH1(R132H) mutation increases murine haematopoietic progenitors and alters epigenetics. Nature 2012;488:656-9. [Crossref] [PubMed]
- Sasaki M, Knobbe CB, Itsumi M, et al. D-2-hydroxyglutarate produced by mutant IDH1 perturbs collagen maturation and basement membrane function. Genes Dev 2012;26:2038-49. [Crossref] [PubMed]
- Rohle D, Popovici-Muller J, Palaskas N, et al. An inhibitor of mutant IDH1 delays growth and promotes differentiation of glioma cells. Science 2013;340:626-30. [Crossref] [PubMed]
- Chowdhury R, Yeoh KK, Tian YM, et al. The oncometabolite 2-hydroxyglutarate inhibits histone lysine demethylases. EMBO Rep 2011;12:463-9. [Crossref] [PubMed]
- Kriaucionis S, Heintz N. The nuclear DNA base 5-hydroxymethylcytosine is present in Purkinje neurons and the brain. Science 2009;324:929-30. [Crossref] [PubMed]
- Tahiliani M, Koh KP, Shen Y, et al. Conversion of 5-methylcytosine to 5-hydroxymethylcytosine in mammalian DNA by MLL partner TET1. Science 2009;324:930-5. [Crossref] [PubMed]
- Xu W, Yang H, Liu Y, et al. Oncometabolite 2-hydroxyglutarate is a competitive inhibitor of alpha-ketoglutarate-dependent dioxygenases. Cancer Cell 2011;19:17-30. [Crossref] [PubMed]
- Yan H, Bigner DD, Velculescu V, et al. Mutant metabolic enzymes are at the origin of gliomas. Cancer Res 2009;69:9157-9. [Crossref] [PubMed]
- Ye D, Xiong Y, Guan KL. The mechanisms of IDH mutations in tumorigenesis. Cell Res 2012;22:1102-4. [Crossref] [PubMed]
- Losman JA, Kaelin WG Jr. What a difference a hydroxyl makes: mutant IDH, (R)-2-hydroxyglutarate, and cancer. Genes Dev 2013;27:836-52. [Crossref] [PubMed]
- Noushmehr H, Weisenberger DJ, Diefes K, et al. Identification of a CpG island methylator phenotype that defines a distinct subgroup of glioma. Cancer Cell 2010;17:510-22. [Crossref] [PubMed]
- Turcan S, Rohle D, Goenka A, et al. IDH1 mutation is sufficient to establish the glioma hypermethylator phenotype. Nature 2012;483:479-83. [Crossref] [PubMed]
- Lu C, Ward PS, Kapoor GS, et al. IDH mutation impairs histone demethylation and results in a block to cell differentiation. Nature 2012;483:474-8. [Crossref] [PubMed]
- Bickmore WA, van Steensel B. Genome architecture: domain organization of interphase chromosomes. Cell 2013;152:1270-84. [Crossref] [PubMed]
- Rao SS, Huntley MH, Durand NC, et al. A 3D map of the human genome at kilobase resolution reveals principles of chromatin looping. Cell 2014;159:1665-80. [Crossref] [PubMed]
- Flavahan WA, Drier Y, Liau BB, et al. Insulator dysfunction and oncogene activation in IDH mutant gliomas. Nature 2016;529:110-4. [Crossref] [PubMed]
- Houillier C, Wang X, Kaloshi G, et al. IDH1 or IDH2 mutations predict longer survival and response to temozolomide in low-grade gliomas. Neurology 2010;75:1560-6. [Crossref] [PubMed]
- SongTao Q, Lei Y, Si G, et al. IDH mutations predict longer survival and response to temozolomide in secondary glioblastoma. Cancer Sci 2012;103:269-73. [Crossref] [PubMed]
- Cairncross JG, Wang M, Jenkins RB, et al. Benefit from procarbazine, lomustine, and vincristine in oligodendroglial tumors is associated with mutation of IDH. J Clin Oncol 2014;32:783-90. [Crossref] [PubMed]
- van den Bent MJ, Brandes AA, Taphoorn MJ, et al. Adjuvant procarbazine, lomustine, and vincristine chemotherapy in newly diagnosed anaplastic oligodendroglioma: long-term follow-up of EORTC brain tumor group study 26951. J Clin Oncol 2013;31:344-50. [Crossref] [PubMed]
- Wakimoto H, Tanaka S, Curry WT, et al. Targetable signaling pathway mutations are associated with malignant phenotype in IDH-mutant gliomas. Clin Cancer Res 2014;20:2898-909. [Crossref] [PubMed]
- Wang F, Travins J, DeLaBarre B, et al. Targeted inhibition of mutant IDH2 in leukemia cells induces cellular differentiation. Science 2013;340:622-6. [Crossref] [PubMed]
- Kernytsky A, Wang F, Hansen E, et al. IDH2 mutation-induced histone and DNA hypermethylation is progressively reversed by small-molecule inhibition. Blood 2015;125:296-303. [Crossref] [PubMed]
- Borodovsky A, Salmasi V, Turcan S, et al. 5-azacytidine reduces methylation, promotes differentiation and induces tumor regression in a patient-derived IDH1 mutant glioma xenograft. Oncotarget 2013;4:1737-47. [Crossref] [PubMed]
- Grassian AR, Parker SJ, Davidson SM, et al. IDH1 mutations alter citric acid cycle metabolism and increase dependence on oxidative mitochondrial metabolism. Cancer Res 2014;74:3317-31. [Crossref] [PubMed]
- Tateishi K, Wakimoto H, Iafrate AJ, et al. Extreme Vulnerability of IDH1 Mutant Cancers to NAD+ Depletion. Cancer Cell 2015;28:773-84. [Crossref] [PubMed]
- Turcan S, Fabius AW, Borodovsky A, et al. Efficient induction of differentiation and growth inhibition in IDH1 mutant glioma cells by the DNMT Inhibitor Decitabine. Oncotarget 2013;4:1729-36. [Crossref] [PubMed]
- Stein EM, Dinardo C, Altman JK, et al. Safety and Efficacy of AG-221, a Potent Inhibitor of Mutant IDH2 That Promotes Differentiation of Myeloid Cells in Patients with Advanced Hematologic Malignancies: Results of a Phase 1/2 Trial. Blood 2015;126:323.
- Burris H, Mellinghoff I, Maher E, et al. The first reported results of AG-120, a first-in-class, potent inhibitor of the IDH1 mutant protein, in a Phase I study of patients with advanced IDH1-mutant solid tumors, including gliomas. Mol Cancer Ther 2015;14:Abstract PL04-5.
- Cruz FD, Matushansky I. Solid tumor differentiation therapy - is it possible? Oncotarget 2012;3:559-67. [Crossref] [PubMed]
- Izquierdo-Garcia JL, Cai LM, Chaumeil MM, et al. Glioma cells with the IDH1 mutation modulate metabolic fractional flux through pyruvate carboxylase. PLoS One 2014;9:e108289. [Crossref] [PubMed]
- Reitman ZJ, Duncan CG, Poteet E, et al. Cancer-associated isocitrate dehydrogenase 1 (IDH1) R132H mutation and d-2-hydroxyglutarate stimulate glutamine metabolism under hypoxia. J Biol Chem 2014;289:23318-28. [Crossref] [PubMed]
- Reitman ZJ, Jin G, Karoly ED, et al. Profiling the effects of isocitrate dehydrogenase 1 and 2 mutations on the cellular metabolome. Proc Natl Acad Sci U S A 2011;108:3270-5. [Crossref] [PubMed]
- Chiarugi A, Dolle C, Felici R, et al. The NAD metabolome--a key determinant of cancer cell biology. Nat Rev Cancer 2012;12:741-52. [Crossref] [PubMed]
- Molenaar RJ, Botman D, Smits MA, et al. Radioprotection of IDH1-Mutated Cancer Cells by the IDH1-Mutant Inhibitor AGI-5198. Cancer Res 2015;75:4790-802. [Crossref] [PubMed]
- Falnes PØ, Johansen RF, Seeberg E. AlkB-mediated oxidative demethylation reverses DNA damage in Escherichia coli. Nature 2002;419:178-82. [Crossref] [PubMed]
- Hegi ME, Diserens AC, Gorlia T, et al. MGMT gene silencing and benefit from temozolomide in glioblastoma. N Engl J Med 2005;352:997-1003. [Crossref] [PubMed]
- Wang P, Wu J, Ma S, et al. Oncometabolite D-2-Hydroxyglutarate Inhibits ALKBH DNA Repair Enzymes and Sensitizes IDH Mutant Cells to Alkylating Agents. Cell Rep 2015;13:2353-61. [Crossref] [PubMed]
- Sulkowski PL, Corso CD, Robinson ND, et al. 2-Hydroxyglutarate produced by neomorphic IDH mutations suppresses homologous recombination and induces PARP inhibitor sensitivity. Sci Transl Med 2017;9. [Crossref] [PubMed]
- Chalmers A, Jackson A, Swaisland H, et al. Results of stage 1 of the oparatic trial: A phase I study of olaparib in combination with temozolomide in patients with relapsed glioblastoma. J Clin Oncol 2014.32. Abstract 2025.
- Muscal JA, Thompson PA, Giranda VL, et al. Plasma and cerebrospinal fluid pharmacokinetics of ABT-888 after oral administration in non-human primates. Cancer Chemother Pharmacol 2010;65:419-25. [Crossref] [PubMed]
- Su JM, Thompson P, Adesina A, et al. A phase I trial of veliparib (ABT-888) and temozolomide in children with recurrent CNS tumors: a pediatric brain tumor consortium report. Neuro Oncol 2014;16:1661-8. [Crossref] [PubMed]
- Tran AN, Lai A, Li S, et al. Increased sensitivity to radiochemotherapy in IDH1 mutant glioblastoma as demonstrated by serial quantitative MR volumetry. Neuro Oncol 2014;16:414-20. [Crossref] [PubMed]
- Cuyàs E, Fernandez-Arroyo S, Corominas-Faja B, et al. Oncometabolic mutation IDH1 R132H confers a metformin-hypersensitive phenotype. Oncotarget 2015;6:12279-96. [Crossref] [PubMed]
- Li F, He X, Ye D, et al. NADP(+)-IDH Mutations Promote Hypersuccinylation that Impairs Mitochondria Respiration and Induces Apoptosis Resistance. Mol Cell 2015;60:661-75. [Crossref] [PubMed]
- Seltzer MJ, Bennett BD, Joshi AD, et al. Inhibition of glutaminase preferentially slows growth of glioma cells with mutant IDH1. Cancer Res 2010;70:8981-7. [Crossref] [PubMed]
- Zhao S, Lin Y, Xu W, et al. Glioma-derived mutations in IDH1 dominantly inhibit IDH1 catalytic activity and induce HIF-1alpha. Science 2009;324:261-5. [Crossref] [PubMed]
- Wang G, Sai K, Gong F, et al. Mutation of isocitrate dehydrogenase 1 induces glioma cell proliferation via nuclear factor-kappaB activation in a hypoxia-inducible factor 1-alpha dependent manner. Mol Med Rep 2014;9:1799-805. [PubMed]
- Carbonneau M. The oncometabolite 2-hydroxyglutarate activates the mTOR signalling pathway. Nat Commun 2016;7:12700. [Crossref] [PubMed]
- Laplante M, Sabatini DM. mTOR signaling in growth control and disease. Cell 2012;149:274-93. [Crossref] [PubMed]
- Schumacher T, Bunse L, Pusch S, et al. A vaccine targeting mutant IDH1 induces antitumour immunity. Nature 2014;512:324-7. [Crossref] [PubMed]
- Pellegatta S, Valletta L, Corbetta C, et al. Effective immuno-targeting of the IDH1 mutation R132H in a murine model of intracranial glioma. Acta Neuropathol Commun 2015;3:4. [Crossref] [PubMed]
- Zhang X, Rao A, Sette P, et al. IDH mutant gliomas escape natural killer cell immune surveillance by downregulation of NKG2D ligand expression. Neuro Oncol 2016;18:1402-12. [Crossref] [PubMed]
- McGilvray RW, Eagle RA, Watson NF, et al. NKG2D ligand expression in human colorectal cancer reveals associations with prognosis and evidence for immunoediting. Clin Cancer Res 2009;15:6993-7002. [Crossref] [PubMed]


