Exploiting molecular genomics in precision radiation oncology: a marriage of biological and physical precision
Introduction
Advances in radiation delivery in the last decade have driven substantial improvement in physical precision of radiotherapy (RT) targeting of tumours. With intensity modulated radiotherapy (IMRT), there is now the ability to shape deposition of radiation doses around concave contours, thereby ensuring dose intensity to the tumour while limiting exposure of adjacent normal tissues. Likewise, image guided radiotherapy (IGRT) with daily cone beam imaging has added another layer of physical precision. Both technologies are culpable for the enhanced therapeutic ratio of RT; clinical evidence from prospective trials and observational audits have indicated significant improvements in disease control, survival, and reduction of normal tissue complications (1,2). More recently, proton therapy has been proposed as an even more superior technology for RT delivery through exploiting the Bragg’s peak characteristics of the proton beam; this yields the theoretical advantage of focused dose intensity at the tumour, while further limiting the exposure to normal tissues due to the sharp dose fall-off. It is through the advent of these advanced technologies that has partly motivated the design of RT fractionation schemes that are intended to achieve “tumour ablation”; these regimes exploit the incremental biological effects of large doses per RT fraction in tumours like prostate and breast cancers that are intrinsically sensitive to fraction size variation (3,4). Clinical evidence supporting the efficacy of such stereotactic ablative (radiosurgical) regimes is being presented by several accompanying reviews in this Chinese Journal of Clinical Oncology Special Issue.
Nonetheless, against the background of technological advances, it is arguable that recent clinical gains from improved physical precision have been less impressive. For example, comparisons of clinical outcomes of patient cohorts who had been treated with either proton therapy or IMRT have not been conclusively in favour of protons (5-8). It is therefore plausible to consider that subsequent therapeutic strategies will require a marriage of physical and biological precision in order to achieve the next substantial gain in precision radiation oncology. It is also coincidental that simultaneous with the progress in RT technologies, there are now high throughput next generation sequencing (NGS) techniques that have improved the efficiency and quality of molecular profiling of human tissues. This has led to the generation of robust datasets on molecular taxonomies of several human cancers by large collaborative consortiums like the International Cancer Genome Consortium (ICGC) and The Cancer Genome Atlas (TCGA) (9-12). Apart from providing a molecular taxonomy on individual cancers (thereby prompting the revisit of our conventional approach of clinical stratification by the TNM stage classification system), other insights gained from these molecular profiling studies include discovery of novel driver mutations that are biomarkers of adverse prognosis and therapeutic resistance. The latter would imply a predictive biomarker that is potentially ‘druggable’ for synergism with standard therapies to counter clonal resistance and tumour recurrence (13,14). This review thus aims to discuss the scientific rationales and considerations underpinning various precision RT strategies, with specific focus on head and neck and prostate cancers. Here, we will first review the current literature on the molecular landscape of these cancers; next, we will discuss the clinical relevance of the molecular features; finally, we will suggest possible approaches of incorporating matched tumour and germline molecular profiling in designing precision RT strategies in the treatment of these cancers.
Search strategy
We searched the PubMed and MEDLINE databases for articles published in English from 01 January 2000 to 30 December 2016 with the keywords ‘molecular’, ‘genomics’, ‘epigenomics’, ‘biomarkers’, ‘prognostic’, ‘predictive’, ‘radiation’, ‘radioresistance’, ‘head and neck’, ‘prostate’, ‘cancers’, ‘carcinoma’, ‘radiotherapy’, ‘radiosurgery’, ‘stereotactic body radiotherapy’, and ‘ablative therapy’. Articles were selected based on relevance, with priority given to highly-cited articles, and articles written in English. Conference abstracts were also reviewed, and considered if they reported statistical methods and hazard ratios (HR), with corresponding confidence intervals (CI) and P values. Articles that preceded the search time-frame were also included if they were highly regarded seminal work.
Molecular characterisation of head and neck and prostate cancers
Epidermal growth factor receptor (EGFR) amplification was among the initial oncogenic driver events that were characterised in head and neck squamous cell cancer (HNSCC) (15-17). As shown by Ang et al. and several others, overexpression of EGFR was common in these tumours, and importantly, predicted for inferior locoregional tumour control following primary RT. Targeting of this receptor through anti-EGFR antibody (cetuximab) was able to inhibit the molecular processes contributing to tumour aggression, thereby improving clinical outcomes in patients treated with combination cetuximab-RT (18). However, More recently there is an emergence of a new phenotype of HNSCC that is associated with human papillomavirus (HPV) infection (19-21). From large-scale epidemiological studies, it was observed that while different HPV-serotypes (-16, -18, and others) have been implicated in head and neck carcinogenesis, oncogenic potential differed between the serotypes, and there is a tropism of HPV to the oropharynx, albeit HPV has also been detected in other head and neck anatomical sub-sites. Of note, the association is strongest between HPV-16 infection and the onset of oropharynx squamous cell carcinoma (OPSCC); it is also in HPV+ OPSCC where these patients have a distinctly more favourable prognosis compared to HPV+ HNSCC of other sites and HPV− HNSCC (22,23). While the oncogenic potential of HPV-16 can be attributed to the expression of E6 and E7 proteins, which are known to inhibit TP53 and RB1, respectively, the mechanistic bases underpinning the favourable prognosis and optimal response of HPV+ OPSCC to RT are less definitive. Disruption of cell cycle checkpoint mechanisms, DNA repair and damage responses have been proposed to account for the increased radiosensitivity of these tumours (24). In the landmark report by TCGA on 279 HNSCCs, it was observed that specific mutational events may be enriched in HPV+ and HPV− HNSCC (activated PIK3CA (56% vs. 34%) and FGFR3 (11% vs. 2%), loss of TRAF3 (22% vs. 1%) in HPV+ tumours; EGFR, FGFR1, CCND1, CDK6, MYC, IGF1R, FAT1 in HPV− tumours). However, these events alone do not fully explain the discordant natural histories of these tumours (12). Other common events such as inactivating mutations in NOTCH1 were also observed, regardless of HPV status, which hints at a novel function of NOTCH1 as a tumour suppressor rather than an oncogene in HNSCC (12,25,26). From a broader genome-wide perspective, intra-tumoral and inter-individual molecular heterogeneity of HNSCC were also appreciable from these landmark sequencing studies, and add prognostic information to conventional clinical indices (27).
Unlike HNSCC, the majority of localised prostate cancers harbour a paucity of somatic nucleotide variants (SNVs); in an analysis of 200 whole genome shotgu (WGS) and 277 whole exome sequences (WES), Fraser et al. reported SNV frequencies of less than 10% for SPOP, MED12, TP53, FOXA1 in non-indolent prostate cancers with similar clinical risk profiles (28). Rather, the molecular taxonomy of localised prostate cancers is better defined by copy number aberrations (CNA), structural chromosomal hypermutations (chromothripsis and kataegis) and rearrangements. In particular, Lalonde et al. showed that localised prostate cancers could be classified by global CNA and gene rearrangement profiles into four subclasses, independent of Gleason’s score (GS) (29). The significant intra-tumoral spatial genomic heterogeneity further highlights the biological complexity of multifocal prostate cancers that could not be discerned by conventional GS and histomorphology (30,31). Beyond the variation in global CNA profiles, prostate cancers are also prone to recurrent gene-specific CNAs such as amplification in CMYC, and losses in PTEN, RB1, TP53, CHD1, CDH1, and NKX3-1 (32). Among recurrent gene rearrangements, TMPRSS2: ERG somatic fusion is most common, occurring in 50% of prostate cancers. Nonetheless, novel inversions also have been reported; Fraser et al. described a recurrent inversion at the PTEN gene locus affecting downstream gene expression (28). In a separate cohort of NCCN-defined high-risk advanced tumours, Baca et al. presented a novel concept of “chromoplexy”, whereby inter-dependent DNA translocations and deletions occur to dysregulate prostate cancer genes in a coordinated manner (33). Finally, the tumour microenvironment within the prostate gland is also prone to effects of hypoxia and co-occurrence of subpathologies such as intraductal and cribriform variants, which raises the possibility of interdependency between the intrinsic tumour molecular characteristics and the surrounding microenvironment (34,35). Taken together, the findings of these studies have provided us with a profound insight on several novel biology in these and other human cancers, in hope that we can eventually link these molecular indices to tumour aggression and therapeutic resistance.
Clinical relevance of somatic and germline mutations in HNSCC and prostate cancers
Treatment recommendation of HNSCC is largely based on a risk-adapted approach using conventional clinical indices; early stage disease (TNM stages I and II) is typically managed with either surgery or RT, while locally advanced tumours (TNM stages III and IV) are treated with either definitive chemoRT or surgery followed by adjuvant therapies (RT or chemoRT). Nonetheless, clinical stratification for personalised treatment strategies can be achieved with incorporation of molecular indices. An illustrative example will be overexpression of EGFR and HER2, which are known adverse prognostic biomarkers in this disease, and targeted therapeutics such as cetuximab and gefitinib are effective in “drugging” this activated pathway in HNSCC. Although modest response rates of 15–20% to either agents in unselected patient populations have been reported (36,37), presence of a germline mutation in KRAS may be predictive of cetuximab efficacy [HR for overall survival (OS) =0.19, P=0.03; progression-free survival (PFS) =0.31, P=0.04 in favour of KRAS-variant] (38). Separately, tumours eventually acquire resistance to these agents through co-occurrence of other gene alterations in ERBB2, MET, PIK3CA, PTEN and HRAS (12). Detailed characterisation of molecular pathways that are aberrant prior to and at the point of resistance would thus provide the scientific rationales of novel combinatorial targeted therapeutic regimes.
Assessment of HPV status by immunohistochemical staining of its surrogate marker—p16 and/or HPV RNA in situ hybridisation (ISH) is routine at present when performing histopathological analysis of HNSCCs, even though information on prognosis is primarily limited to OPSCC (23). Bratman et al. further suggested that prognostication power varies between HPV-serotypes; in their analysis of 73 HPV+ tumours from the TCGA cohort, they observed that HPV-16 was associated with superior survival when compared to HPV-other serotypes (39). Nonetheless, it is conclusive that the strength of HPV as a prognostic biomarker far outweighs conventional clinical indices in HNSCC; of note, a novel stage classification system that is unique to HPV+ OPSCC has thus been developed (40,41). The enhanced radiation sensitivity of these viral-associated tumours has also led to design of treatment deintensification regimes, with preliminary evidence from few phase II clinical trials supportive of a less intensive approach (42-44). Separately, the preponderance of distinct mutations like activated PIK3CA in HPV+ OPSCC lends itself to the possibility of exploiting tumour susceptibility to inhibitors of PIK3CA (buparlisib, alpelisib, etc.).
Likewise in prostate cancers, patient stratification using conventional clinical indices alone (CT category, GS, and pre-treatment PSA) is imprecise, with 30% of patients failing primary surgery or RT (45). On this note, genomic instability, CMYC gain, TP53/RB1 deletion are among the molecular aberrations that are associated with risks of biochemical and metastatic relapses following definitive surgery or RT (29,46-48). Dysregulation of non-coding genes has also been proposed to portend for unfavourable prognoses; expression of a long non-coding RNA, SChLAP1, which is specifically expressed in prostate tissue, has been described in lethal prostate cancers (49,50).
Apart from providing information on lethality of these cancers, mutations such as somatic NBN amplification and germline HSD3B1 activating variants have been separately reported to predict for RT-resistance [5-year biochemical relapse-free rate (bRFR) of 46% (gain) vs. 77% (no gain), P=0.00067 in IGRT cohort; no difference in prostatectomy cohort] and insensitivity to androgen suppression (median PFS of 6.6-year in homozygous wild type vs. 4.1-year in heterozygous variants vs. 2.5-year in homozygous variants, P=0.011), respectively (51); these biomarkers have potential utility in refining treatment recommendations in patients who have otherwise similar clinical risk profiles. In the same vein, Zhao et al. performed unsupervised hierarchical clustering of transcriptomes from 3,782 prostate cancers, and observed similar profiles akin to oestrogen-sensitive breast cancers: luminal A, luminal B, and basal (52). However, in contrast luminal B signature conferred the worst prognoses in prostate cancer when compared to basal and luminal A subtypes (10-year bRFR of 29% vs. 39% vs. 41%; 10-year distant metastasis-free survival =53% vs. 73% vs. 73%; 10-year prostate cancer-specific survival =76% vs. 86% vs. 89%, respectively); this is posited to be due to intrinsic insensitivity to androgen suppression in luminal B tumours.
More recently, mutations in DNA repair genes have also been implicated in the development of aggressive prostate cancer and response to targeted therapeutics; a prime example is the anti-tumour activity with inhibition of PARP1 in metastatic prostate cancers harbouring germline and somatic mutations in DNA repair genes, particularly in those involved in homologous recombination (53,54). Interestingly, germline mutations in DNA repair genes were also more frequent among patients with aggressive metastatic disease compared to their indolent counterpart (11.8% vs. 4.6%), with BRCA2 (5.3%) being the most frequent aberration (54). These findings justify the approach of molecular profiling of paired prostate tumour-normal tissues, in order to determine the optimal therapeutic strategy for the individual patient (Table 1) (55-65).
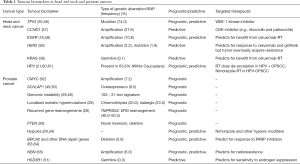
Full table
Optimising RT therapeutic ratio: germline genetic predictors of normal tissue toxicities
Apart from inter-individual heterogeneity in tumour response to RT, a wide variation also exists in normal tissue RT-response between non-syndromic individuals, which are unexplained by clinical and treatment parameters (66,67). It is estimated that up to 60% of inter-individual heterogeneity in normal tissue RT-sensitivity may be due to intrinsic genetic susceptibility, likely influenced by low penetrance allelic variation (68). Identifying genetic signatures of normal tissue RT-sensitivity could therefore offer another paradigm of precision RT; de-escalating RT doses in individuals assessed to be RT-sensitive (by signature) and intensifying treatment in patients who are determined to be less prone to RT-induced normal tissue toxicities. There are now several genome-wide association studies (GWAS) that have investigated for predictive single nucleotide polymorphism (SNP)-based signatures of specific RT-induced normal tissue end-points: Radiogenomics: Assessment of Polymorphisms for Predicting the Effects of Radiotherapy (RAPPER), RADIOGEN, Gene-PARE and Cross Cancer Institute (CCI) (69-73) (Table 2); notably, SNPs in TANC1 and XRCC1 have been reported to predict for erectile dysfunction and breast fibrosis, respectively (71,72). While we await validation in larger cohorts (73,74), other assays including radiation-induced lymphocyte apoptosis (RILA) have been prospectively validated to predict for severe reactions following breast RT (75,76). Going forward, it is plausible to consider a clinical pathway that personalises RT doses for each patient based on profiling data of matched tumour and normal tissue RT-sensitivity in the same patient.

Full table
Designing biomarker-directed therapeutic strategies in head and neck and prostate cancers
Given the RT-sensitivity and favourable prognosis of HPV+ OPSCC, the feasibility of RT dose de-escalation were formally tested in small single-arm phase II studies, with encouraging preliminary results (77). In two of the studies that have been reported, response to induction chemotherapy was employed as a method of stratifying patients to a lower RT dose of 54 Gy, while simultaneously targeting occult metastases with the upfront administration of systemic therapy (43,44). While the systemic therapy regimes varied in both studies (three cycles of induction cisplatin, paclitaxel, cetuximab and concurrent cetuximab in ECOG 1308; two cycles of induction carboplatin, paclitaxel and concurrent paclitaxel in the UCLA trial), the study investigators reported 2-year PFS of 80% and 92%, respectively.
Treatment intensification strategies in HPV− HNSCC have included testing the efficacy of combination hypoxia modifiers (nimorazole, tirapazamine) and RT, with the aim of sensitising radioresistant hypoxic clones in these tumours. The background to this strategy was in part based on a post-hoc analysis of p16 expression as a surrogate for HPV-status in the DAHANCA-5 randomised controlled phase III trial of nimorazole-RT compared to RT alone (78); the study investigators observed that negative expression of p16 was predictive of nimorazole efficacy (HR of loco-regional failure with nimorazole 0.69, p16- vs. 0.93, p16+) (79). This strategy is currently being formally investigated in a randomised controlled phase III trial (ClinicalTrials.gov, NCT01880359) (80). Another rationale for treatment intensification in this adverse subgroup involves the targeting of occult metastases. To this end, investigators have examined the role of maintenance afatinib (an EGFR and HER2 tyrosine kinase inhibitor) following chemo-RT in two large randomised studies (LUX-2 and LUX-4), both of which unfortunately failed to demonstrate an efficacy of the targeted agent in this setting (81). Novel regimes are therefore needed, and we await the results of ongoing immunotherapy trials [ClinicalTrials.gov, NCT02952586 (JAVELIN), NCT02777385, NCT02764593 (RTOG 3504)].
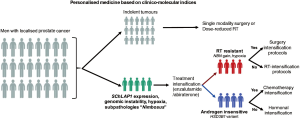
A similar approach could also be applied in the treatment of localised prostate cancer; while most tumours are indolent in nature, and could be considered for single modality surgery or RT without dose escalation, those harbouring aggressive features of SChLAP1 expression and genomic instability ought to be considered for treatment intensification with second generation anti-androgen therapies (enzalutamide and abiraterone). In addition, biomarkers that are predictive of RT- (NBN amplification) and androgen-sensitivity (HSD3B1 variant) could also be incorporated into a clinical decision making tool comprising of combinatorial clinical indices and prognostic biomarkers; for example, in individuals with prostate tumours harbouring a constellation of hypoxia, subpathologies, and genomic instability (46), these men should be referred for treatment intensification clinical trial protocols, which could embed additional stratification measures to (I) surgery or RT based on an RT-sensitivity signature, and (II) chemotherapy or hormonal therapy for targeting systemic disease depending on their germline HSD3B1 status (Figure 1). In the metastatic setting, germline and tumour sequencing should be performed, since this could help direct patients to treatment with PARP1 inhibitors (olaparib, etc.) in the presence of mutations in DNA repair genes (53).
Nonetheless, there are several caveats with such precision approaches. Foremost, large-scale prospective validation is required for several of these molecular biomarkers to verify the magnitude of association with clinical outcomes. Next, standardisation of the molecular assays to achieve robust reproducibility and reliability is paramount to ensuring consistency of the assay read-out across multiple centres. Lastly, a challenge yet to be adequately addressed relates to the impact of intratumoral heterogeneity on the accuracy of genotype-phenotype (82), and if liquid biopsies represent the solution to more invasive and costlier multiregional tumour sampling.
Combinatorial approach of precision molecular profiling and ablative stereotactic RT
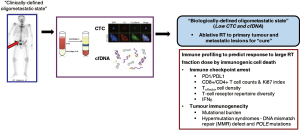
While the role of stereotactic body RT (SBRT) or radiosurgery (SRS) was initially intended for optimisation of symptom palliation, there is an emerging concept among clinicians to exploit SBRT and SRS in targeting isolated metastases, such that these patients harbouring a “low” burden of systemic metastases are potentially “cured” if the metastatic tumour clones are eradicated. Supportive evidence for such a disease state can be drawn from clinical examples in colorectal, renal, and soft tissue cancers (83). Nonetheless, current methods of stratifying for these favourable patients are imprecise, and do not incorporate indices indicative of tumour biology; they often rely on assessment for (I) number of metastatic lesions; (II) sites of metastasis; (III) disease-free interval; and (IV) control of primary disease. In this instance, one could identify the relevance of a molecular signature for an oligometastatic state, which would enhance our ability to identify patients with truly limited disease. Of note, liquid biopsies that rely on quantification and characterisation of circulating tumour cells (CTCs) and cell-free tumour DNA (cfDNA) could fulfil this clinical purpose, and have demonstrated preliminary success in early-stage colorectal and lung cancers (84,85). In the larger TRAcking Cancer Evolution through therapy (Rx) (TRACERx) study, Abbosh et al. reported on a novel technology that could detect cfDNA in 100 early-stage lung cancer patients, coupled with a high fidelity in phylogenetic analyses to track clonal evolution that could predict resistance to adjuvant chemotherapy and risk of recurrence (84). Separately, Tie and colleagues demonstrated that detection of tumour-associated mutations in cfDNA was predictive of a benefit with adjuvant chemotherapy in patients with stage 2 colorectal cancer following surgery (85). Based on these examples, it is plausible to consider the utility of this technology for a more accurate stratification of patients with oligometastatic disease, who would be ideal candidates for more aggressive intervention.
Precision molecular profiling could also be useful for evaluating tumour immunogenicity. As discussed elegantly in the companion review article by Tharmalingam and Hoskin (86), there is good level of evidence that innate and adaptive anti-tumour immune responses are triggered by RT, and RT-induced immunogenic cell death (ICD) is particularly dominant at large fraction sizes (87). Moreover, the synergism that is observed with combining RT and immune checkpoint inhibitors [anti-programmed cell death-1 (PD1) and ligand-1 (PDL-1)], anti-cytotoxic T lymphocyte-associated protein-4 (CTLA4) supports ICD as a main mechanism of SBRT and SRS anti-tumour efficacy (88). ICD can be exerted through an acute release of tumour-associated antigens release leading to priming of cytotoxic T-cells, and recruitment of antigen presenting cells (APC) and dendritic cells (89). Therefore, profiling of tumour mutational burden and the immune microenvironment ought to be performed in ongoing trials of combination immuno-RT, in order to derive potential predictive signatures. The above outlined concepts are illustrated in Figure 2.
Conclusions
In the past decade, the radiation oncology community has embraced the technological advances that have transitioned the field into an era of precision RT, which have not only contributed to tremendous gains in tumour control probability and reduction of normal tissue toxicities, but in the same vein, catered for novel tumour ablative treatments. Consequently, we have witnessed some practice-changing transitions in cancer management; examples include the safe delivery of prostate RT in five fractions over two weeks as opposed to two months of conventionally fractionated RT, and the ablation of oligometastasis resulting in curing of patients with disseminated disease. These are landmark improvements in patient survivorship and quality of life. That said, we argue that the next wave of therapeutic gains will come from biological precision strategies. Here, we presented data on our current understanding into the molecular drivers of carcinogenesis, tumour aggression, and treatment resistance in HNSCC and prostate cancer. We reported on how some of these molecular indices have the potential utility in patient prognostication and influencing treatment recommendation. As we deepen our scientific understanding, it is only imaginable that individualised prescription of RT doses and combination regimes will become a reality in due course.
Acknowledgements
The authors thank all members of the Chua and Soo laboratories for support and guidance.
Funding: ML Chua is supported by the National Medical Research Council Singapore Transition Award (NMRC/TA/0030/2014) and the Duke-NUS Oncology Academic Program Proton Therapy Research Program. X Lin and KC Soo are supported by the National Medical Research Council grants (NMRC/CIRG/1355/2013, NMRC/CBRG/0068/2014) and SingHealth Foundation grant (SHF/FG573P/2014). PY Lam is supported by the NMRC Centre grant.
Footnote
Conflicts of Interest: The authors have no conflicts of interest to declare.
References
- Buettner F, Miah AB, Gulliford SL, et al. Novel approaches to improve the therapeutic index of head and neck radiotherapy: an analysis of data from the PARSPORT randomised phase III trial. Radiother Oncol 2012;103:82-7. [Crossref] [PubMed]
- Nutting CM, Mordern JP, Harrington KJ, et al. Parotid-sparing intensity modulated versus conventional radiotherapy in head and neck cancer (PARSPORT): a phase 3 multicentre randomised controlled trial. Lancet Oncol 2011;12:127-36. [Crossref] [PubMed]
- Yarnold J, Ashton A, Bliss J, et al. Fractionation sensitivity and dose response of late adverse effects in the breast after radiotherapy for early breast cancer: long term results of a randomised trial. Radiother Oncol 2005;75:9-17. [Crossref] [PubMed]
- Somaiah N, Yarnold J, Daley F, et al. The relationship between homologous recombination repair and the sensitivity of human epidermis to the size of daily doses over a 5-week course of breast radiotherapy. Clin Cancer Res 2012;18:5479-88. [Crossref] [PubMed]
- Hoppe BS, Michalski JM, Mendenhall NP, et al. Comparative effectiveness study of patient-reported outcomes after proton therapy or intensity-modulated radiotherapy for prostate cancer. Cancer 2014;120:1076-82. [Crossref] [PubMed]
- Blanchard P, Garden AS, Gunn GB, et al. Intensity-modulated proton beam therapy (IMPT) versus intensity-modulated photon therapy (IMRT) for patients with oropharynx cancer - A case matched analysis. Radiother Oncol 2016;120:48-55. [Crossref] [PubMed]
- Yamoah K, Johnstone PA. Proton beam therapy: clinical utility and current status in prostate cancer. Onco Targets Ther 2016;9:5721-7. [Crossref] [PubMed]
- Liao ZX, Lee J, Komaki R, et al. Bayesian randomised trial comparing intensity modulated radiotherapy versus passively scattered proton therapy for locally advanced non-small cell lung cancer. J Clin Oncol 2016;34:8500.
- International Cancer Genome Consortium, Hudson TJ, Anderson W, et al. International network of cancer genome projects. Nature 2010;464:993-8. [Crossref] [PubMed]
- Song Y, Li L, Ou Y, et al. Identification of genomic alterations in oesophageal squamous cell cancer. Nature 2014;509:91-5. [Crossref] [PubMed]
- The Cancer Genome Atlas Research Network. Comprehensive molecular profiling of lung adenocarcinoma. Nature 2014;511:543-50. [Crossref]
- The Cancer Genome Atlas Network. Comprehensive genomic characterization of head and neck squamous cell carcinomas. Nature 2015;517:576-82. [Crossref] [PubMed]
- Chin L, Andersen JN, Futreal PA. Cancer genomics: from discovery science to personalised medicine. Nat Med 2011;17:297-303. [Crossref] [PubMed]
- Boutros PC, Fraser M, van der Kwast T, et al. Clonality of localized and metastatic prostate cancer. Curr Opin Urol 2016;26:219-24. [Crossref] [PubMed]
- Dassonville O, Formento JL, Francoual M, et al. Expression of epidermal growth factor receptor and survival in upper aerodigestive tract cancer. J Clin Oncol 1993;11:1873-8. [Crossref] [PubMed]
- Rubin Grandis J, Melhem MF, Gooding WE, et al. Levels of TGF-alpha and EGFR protein in head and neck squamous cell carcinoma and patient survival. J Natl Cancer Inst 1998;90:824-32. [Crossref] [PubMed]
- Ang KK, Andratschke NH, Milas L. Epidermal growth factor receptor and response of head-and-neck carcinoma to therapy. Int J Radiat Oncol Biol Phys 2004;58:959-65. [Crossref] [PubMed]
- Bonner JA, Harari PM, Giralt J, et al. Radiotherapy plus cetuximab for squamous-cell carcinoma of the head and neck. N Engl J Med 2006;354:567-78. [Crossref] [PubMed]
- Gillison ML, Koch WM, Capone RB, et al. Evidence for a causal association between human papillomavirus and a subset of head and neck cancers. J Natl Cancer Inst 2000;92:709-20. [Crossref] [PubMed]
- Mork J, Lie AK, Glattre E, et al. Human papillomavirus infection as a risk factor for squamous-cell carcinoma of the head and neck. N Engl J Med 2001;344:1125-31. [Crossref] [PubMed]
- Ang KK, Harris J, Wheeler R, et al. Human papillomavirus and survival of patients with oropharyngeal cancer. N Engl J Med 2010;363:24-35. [Crossref] [PubMed]
- Nguyen-Tan PF, Zhang Q, Ang KK, et al. Randomised phase III trial to test accelerated versus standard fractionation in combination with concurrent cisplatin for head and neck carcinomas in the Radiation Therapy Oncology Group 0129 trial: long term report of efficacy and toxicity. J Clin Oncol 2014;32:3858-66. [Crossref] [PubMed]
- Posner MR, Lorch JH, Goloubeva O, et al. Survival and human papillomavirus in oropharynx cancer in TAX 324: a subset analysis from an international phase III trial. Ann Oncol 2011;22:1071-7. [Crossref] [PubMed]
- Wang L, Zhang P, Molkentine DP, et al. TRIP12 as a mediator of human papillomavirus/p16-related radiation enhancement effects. Oncogene 2017;36:820-8. [Crossref] [PubMed]
- Agrawal N, Frederick MJ, Pickering CR, et al. Exome sequencing of head and neck squamous cell carcinoma reveals inactivating mutations in NOTCH1. Science 2011;333:1154-7. [Crossref] [PubMed]
- Stransky N, Egloff AM, Tward AD, et al. The mutational landscape of head and neck squamous cell carcinoma. Science 2011;333:1157-60. [Crossref] [PubMed]
- Mroz EA, Tward AD, Hammon RJ, et al. Intratumor genetic heterogeneity and mortality in head and neck cancer: analysis of data from the Cancer Genome Atlas. PLoS Med 2015;12:e1001818. [Crossref] [PubMed]
- Fraser M, Sabelnykova VY, Takafumi N, et al. Genomic hallmarks of localized, non-indolent prostate cancer. Nature 2017;541:359-64. [Crossref] [PubMed]
- Lalonde E, Ishkanian AS, Sykes J, et al. Tumour genomic and microenvironmental heterogeneity for integrated prediction of 5-year biochemical recurrence of prostate cancer: a retrospective cohort study. Lancet Oncol 2014;15:1521-32. [Crossref] [PubMed]
- Boutros PC, Fraser M, Harding NJ, et al. Spatial genomic heterogeneity within localized, multifocal prostate cancer. Nat Genet 2015;47:736-45. [Crossref] [PubMed]
- Cooper CS, Eeles R, Wedge DC, et al. Analysis of the genetic phylogeny of multifocal prostate cancer identifies multiple independent clonal expansions in neoplastic and morphologically normal prostate tissue. Nat Genet 2015;47:367-72. [Crossref] [PubMed]
- Fraser M, Van der Kwast T, Boutros PC, et al. The clinical genomics of prostate cancer. In: Bolla M, van Poppel H. editors. Management of prostate cancer. Switzerland: Springer International Publishing, 2017.
- Baca SC, Prandi D, Lawrence MS, et al. Punctuated evolution of prostate cancer genomes. Cell 2013;153:666-77. [Crossref] [PubMed]
- Milosevic M, Warde P, Menard C, et al. Tumor hypoxia predicts biochemical failure following radiotherapy for clinically localized prostate cancer. Clin Cancer Res 2012;18:2108-14. [Crossref] [PubMed]
- Parker C, Milosevic M, Toi A, et al. Polarographic electrode study of tumor oxygenation in clinically localized prostate cancer. Int J Radiat Oncol Biol Phys 2004;58:750-7. [Crossref] [PubMed]
- Vermorken JB, Trigo J, Hitt R, et al. Open-label, uncontrolled, multicentre phase II study to evaluate the efficacy and toxicity of cetuximab as a single agent in patients with recurrent and/or metastatic squamous cell carcinoma of the head and neck who failed to respond to platinum based therapy. J Clin Oncol 2007;25:2171-7. [Crossref] [PubMed]
- Machiels JP, Haddad R, Fayette J, et al. Afatinib versus methotrexate as second-line treatment in patients with recurrent or metastatic squamous-cell carcinoma of the head and neck progressing on or after platinum-based therapy (LUX-Head & Neck 1): an open-label, randomised phase 3 trial. Lancet Oncol 2015;16:583-94. [Crossref] [PubMed]
- Weidhaas JB, Harris J, Schaue D, et al. The KRAS-variant and cetuximab response in head and neck squamous cell cancer. JAMA Oncol 2017;3:483-91. [Crossref] [PubMed]
- Bratman SV, Bruce JP, O’Sullivan B, et al. Human papillomavirus genotype association with survival in head and neck squamous cell carcinoma. JAMA Oncol 2016;2:823-26. [Crossref] [PubMed]
- Huang SH, Xu W, Waldron J, et al. Refining American Joint Committee on Cancer/Union for International Cancer Control TNM stage and prognostic groups for human papillomavirus-related oropharyngeal carcinomas. J Clin Oncol 2015;33:836-45. [Crossref] [PubMed]
- O'Sullivan B, Huang SH, Su J, et al. Development and validation of a staging system for HPV-related oropharyngeal cancer by the International Collaboration on Oropharyngeal cancer Network for Staging (ICON-S): a multicentre cohort study. Lancet Oncol 2016;17:440-51. [Crossref] [PubMed]
- Chera BS, Amdur RJ, Tepper J, et al. Phase 2 trial of De-intensified chemoradiation therapy for favourable-risk human papillomavirus-associated oropharyngeal squamous cell carcinoma. Int J Radiat Oncol Biol Phys 2015;93:976-85. [Crossref] [PubMed]
- Marur S, Li S, Cmelak AJ, et al. E1308: phase II trial of induction chemotherapy followed by reduced-dose radiation and weekly cetuximab in patients with HPV-associated resectable squamous cell carcinoma of the oropharynx- ECOG-ACRIN Cancer Research Group. J Clin Oncol 2016. [Epub ahead of print]. [PubMed]
- Chen AM, Felix C, Wang PC, et al. Reduced-dose radiotherapy for human papillomavirus-associated squamous-cell carcinoma of the oropharynx: a single-arm, phase 2 study. Lancet Oncol 2017;18:803-11. [Crossref] [PubMed]
- D’Amico AV, Moul J, Carroll PR, et al. Cancer specific mortality after surgery or radiation for patients with clinically localized prostate cancer managed during the prostate-specific antigen era. J Clin Oncol 2003;21:2163-72. [Crossref] [PubMed]
- Chua MLK, Lo W, Pintilie M, et al. A prostate cancer "nimbosus": genomic instability and SChLAP1 dysregulation underpin aggression of intraductal and cribriform subpathologies. Eur Urol 2017. [Epub ahead of print]. [Crossref] [PubMed]
- Ku SY, Spencer R, Wang YQ, et al. Rb1 and Trp53 cooperate to suppress prostate cancer lineage plasticity, metastasis, and antiandrogen resistance. Science 2017;355:78-83. [Crossref] [PubMed]
- Lalonde E, Alkallas R, Chua MLK, et al. Translating a prognostic DNA genomic classifier into the clinic: retrospective validation in 563 localized prostate tumors. Eur Urol 2017;72:22-31. [Crossref] [PubMed]
- Presner JR, Iyer MK, Sahu A, et al. The long non-coding RNA SChLAP1 promotes aggressive prostate cancer and antagonizes the SWI/SNF complex. Nat Gen 2013;45:1392-98. [Crossref]
- Mehra R, Udager AM, Ahearn TU, et al. Overexpression of the long non-coding RNA SChLAP1 independently predicts lethal prostate cancer. Eur Urol 2016;70:549-52. [Crossref] [PubMed]
- Hearn JW, AbuAli G, Reichard CA, et al. HSD3B1 and resistance to androgen-deprivation therapy in prostate cancer: a retrospective multicohort study. Lancet Oncol 2016;17:1435-44. [Crossref] [PubMed]
- Zhao SG, Chang SL, Erho N, et al. Associations of luminal and basal subtyping of prostate cancer with prognosis and response to androgen deprivation therapy. JAMA Oncol 2017. [Epub ahead of print]. [Crossref] [PubMed]
- Mateo J, Carreira S, Sandhu S, et al. DNA-repair defects and olaparib in metastatic prostate cancer. N Engl J Med 2015;373:1697-708. [Crossref] [PubMed]
- Pritchard CC, Mateo J, Walsh MF, et al. Inherited DNA-repair gene mutations in men with metastatic prostate cancer. N Engl J Med 2016;375:443-53. [Crossref] [PubMed]
- Poeta ML, Manola J, Goldwasser MA, et al. TP53 mutations and survival in squamous-cell carcinoma of the head and neck. N Engl J Med 2007;357:2552-61. [Crossref] [PubMed]
- Moser R, Xu C, Kao M, et al. Functional kinomics identifies candidate therapeutic targets in head and neck cancer. Clin Cancer Res 2014;20:4274-88. [Crossref] [PubMed]
- Aung KL, Siu LL. Genomically personalized therapy in head and neck cancer. Cancers Head Neck 2016;1:2. [Crossref]
- Ang KK, Berkey BA, Tu X, et al. Impact of epidermal growth factor receptor expression on survival and pattern of relapse in patients with advanced head and neck carcinoma. Cancer Res 2002;62:7350-6. [PubMed]
- Pollock NI, Grandis JR. HER2 as a therapeutic target in head and neck squamous cell carcinoma. Clin Cancer Res 2015;21:526-33. [Crossref] [PubMed]
- Fakhry C, Westra WH, Li S, et al. Improved survival of patients with human papillomavirus-positive head and neck squamous cell carcinoma in a prospective clinical trial. J Natl Cancer Inst 2008;100:261-9. [Crossref] [PubMed]
- Lassen P, Overgaard J, Eriksen JG. Expression of EGFR and HPV-associated p16 in oropharyngeal carcinoma: correlation and influence on prognosis after radiotherapy in the randomized DAHANCA 5 and 7 trials. Radiother Oncol 2013;108:489-94. [Crossref] [PubMed]
- Zeng W, Sun H, Meng F, et al. Nuclear C-MYC expression level is associated with disease progression and potentially predictive of two year overall survival in prostate cancer. Int J Clin Exp Pathol 2015;8:1878-88. [PubMed]
- Edwards SM, Evans DG, Hope Q, et al. Prostate cancer in BRCA2 germline mutation carriers is associated with poorer prognosis. Br J Cancer 2010;103:918-24. [Crossref] [PubMed]
- Sandhu SK, Omlin A, Hylands L, et al. Poly (ADP-ribose) polymerase (PARP) I inhibitors for the treatment of advanced germline BRCA2 mutant prostate cancer. Ann Oncol 2013;24:1416-8. [Crossref] [PubMed]
- Berlin A, Lalonde E, Sykes J, et al. NBN gain is predictive for adverse outcome following image-guided radiotherapy for localized prostate cancer. Oncotarget 2014;5:11081-90. [Crossref] [PubMed]
- Turesson I. Individual variation and dose dependency in the progression rate of skin telangiectasia. Int J Radiat Oncol Biol Phys 1990;19:1569-74. [Crossref] [PubMed]
- Chua MLK, Rothkamm K. Biomarkers of radiation exposure: can they predict normal tissue radiosensitivity. Clin Oncol (R Coll Radiol) 2013;25:610-6. [Crossref] [PubMed]
- Barnett GC, Coles CE, Elliott RM, et al. Independent validation of genes and polymorphisms reported to be associated with radiation toxicity: a prospective analysis study. Lancet Oncol 2012;13:65-77. [Crossref] [PubMed]
- Alam A, Mukhopadhyay ND, Ning Y, et al. A preliminary study on racial differences in HMOX1, NFE2L2 and TGFβ1 gene polymorphisms and radiation induced late normal tissue toxicity. Int J Radiat Oncol Biol Phys 2015;93:436-43. [Crossref] [PubMed]
- Alsbeih G, El-Sebaie M, Al-Harbi N, et al. SNPs in genes implicated in radiation response are associated with radiotoxicity and evoke roles as predictive and prognostic biomarkers. Radiat Oncol 2013;8:125. [Crossref] [PubMed]
- Fachal L, Gomez-Caamano A, Barnett GC, et al. A three stage genome-wide association study identifies a susceptibility locus for late radiotherapy toxicity at 2q24.1. Nat Genet 2014;46:891-94. [Crossref] [PubMed]
- Seibold P, Behrens S, Schemezer P, et al. XRCC1 polymorphism associated with late toxicity after radiotherapy in breast cancer patients. Int J Radiat Oncol Biol Phys 2015;92:1084-92. [Crossref] [PubMed]
- Barnett GC, Thompson D, Fachal L, et al. A genome wide association study (GWAS) providing evidence of an association between common genetic variants and late radiotherapy toxicity. Radiother Oncol 2014;111:178-85. [Crossref] [PubMed]
- Kerns SL, Dorling L, Fachal L, et al. Meta-analysis of genome wide association studies identifies genetic markers of late toxicity following radiotherapy for prostate cancer. EBioMedicine 2016;10:150-63. [Crossref] [PubMed]
- Azria D, Belkacemi Y, Romieu G, et al. Concurrent or sequential adjuvant letrozole and radiotherapy after conservative surgery for early-stage breast cancer (CO-HO-RT): a phase 2 randomised trial. Lancet Oncol 2010;11:258-65. [Crossref] [PubMed]
- Azria D, Riou O, Castan F, et al. Radiation-induced CD8 T-lymphocyte apoptosis as a predictor of breast fibrosis after radiotherapy: results of the prospective multicentre French trial. EBioMedicine 2015;2:1965-73. [Crossref] [PubMed]
- Bhatia A, Burtness B. Human papillomavirus-associated oropharyngeal cancer: defining risk groups and clinical trials. J Clin Oncol 2015;33:3243-50. [Crossref] [PubMed]
- Overgaard J, Hansen HS, Overgaard M, et al. A randomized double-blind phase III study of nimorazole as a hypoxic radiosensitizer of primary radiotherapy in supraglottic larynx and pharynx carcinoma. Results of the Danish Head and Neck Cancer Study (DAHANCA) Protocol 5-85. Radiother Oncol 1998;46:135-46. [Crossref] [PubMed]
- Lassen P, Eriksen JG, Hamilton-Dutoit S, et al. HPV- associated p16- expression and response to hypoxic modification of radiotherapy in head and neck cancer. Radiother Oncol 2010;94:30-5. [Crossref] [PubMed]
- Christiaens M, Collette S, Overgaard J, et al. Quality assurance of radiotherapy in the ongoing EORTC 1219-DAHANCA-29 trial for HPV/p16 negative squamous cell carcinoma of the head and neck: Results of the benchmark case procedure. Radiother Oncol 2017;123:424-30. [Crossref] [PubMed]
- Burtness B, Haddad RI, Dinis J, et al. LUX-head and neck 2: randomized, double blind, placebo-controlled, phase III trial of afatinib as adjuvant therapy after chemoradiation (CRT) in primary unresected, high/intermediate-risk, squamous cell cancer of head and neck (HNSCC) patients (pts). J Clin Oncol 2017;35:abstr 6001.
- Wei L, Wang JM, Lampert E, et al. Intratumoral and intertumoral genomic heterogeneity of multifocal localized prostate cancer impacts molecular classifications and genomic prognosticators. Eur Urol 2017;71:183-92. [Crossref] [PubMed]
- Tree AC, Khoo VS, Eeles RA, et al. Stereotactic body radiotherapy for oligometastases. Lancet Oncol 2013;14:e28-37. [Crossref] [PubMed]
- Abbosh C, Birkbak NJ, Wilson GA, et al. Phylogenetic ctDNA analysis depicts early-stage lung cancer evolution. Nature 2017;545:446-51. [Crossref] [PubMed]
- Tie J, Wang Y, Tomasetti C, et al. Circulating tumor DNA analysis detects minimal residual disease and predicts recurrence in patients with stage II colon cancer. Sci Transl Med 2016;8:346ra92. [Crossref] [PubMed]
- Tharmalingam H, Hoskin PJ. The optimism surrounding stereotactic body radiation therapy and immunomodulation. Chinese Clin Oncol 2017;6:S9.
- Qian JM, Yu JB, Kluger HM, et al. Timing and type of immune checkpoint therapy affect the early radiographic response of melanoma brain metastases to stereotactic radiosurgery. Cancer 2016;122:3051-8. [Crossref] [PubMed]
- Twyman-Saint Victor C, Rech AJ, Maity A, et al. Radiation and dual checkpoint blockade activate non-redundant immune mechanisms in cancer. Nature 2015;520:373-77. [Crossref] [PubMed]
- Van Limbergen EJ, De Ruysscher DK, Olivo Pimentel V, et al. Combining radiotherapy with immunotherapy: the past, the present and the future. Br J Radiol 2017;90:20170157. [Crossref] [PubMed]


