
Population diversity in oncology drug responses and implications to drug development
As a result of unravelling of the molecular underpinnings of the cancer phenotype, oncology drug development has proceeded at unprecedented pace, with multiple new entities being approved for use in specific indications. However, most patients who are treated with an approved drug for a specific scenario would not derive clinical benefit from treatment, and be subjected to potential for drug toxicity. Oncology agents often have narrow therapeutic windows, and therefore, to achieve the goal of precision medicine of matching the appropriate drug, dose and schedule with the appropriate patient, it would be necessary to understand and manage sources of variability in drug response. These biomarkers of toxicity and efficacy are valuable to guide drug selection and dosing, thus reducing adverse drug reactions, and improve safety and efficacy of treatments.
Oncology drug development has become increasingly a globalised effort, with the emergence of Asian economy and aging populations creating a need for effective and safe cancer therapeutics. It has been debated for decades whether ethno-geographical variability of drug response is an important scientifically grounded factor to consider during drug development to be considered in the regulatory aspects of drug approval in the various countries. There are recognised differences in geographical demographics of cancers, with certain malignancies like hepatitis B related liver cancer, epidermal growth factor receptor (EGFR) tyrosine kinase (TK) domain mutation-positive lung cancer, Epstein Barr virus-positive nasopharyngeal carcinoma and gastric carcinoma being widely recognised to be more common in certain East Asian countries compared to worldwide averages. Similarly, differences in drug responses, including toxicities to cancer therapeutics have been described between populations that are separated geographically. Drug development in oncology has been traditionally based on establishing a recommended dose that is tolerable and efficacious based on an average effect on a population cohort, typically a Western population cohort. This information on dosing and schedule on the drug label is then extrapolated to the rest of the patient population. Yet the expected population pharmacokinetics and pharmacodynamics of this dose and schedule on an extended population cohort may be variable, and depend on how similar the study cohort is to the population being treated. As we look to individualise therapeutics, it becomes critical to consider how representative the population data is to the individual patient, and to consider this during the drug development phases. This paper intends to raise awareness of the issues and ramifications of defining and addressing population differences in cancer therapeutics, and provide some perspectives on future work that is needed to properly address these issues.
Population variation in drug response
Population differences in cancer pharmacology have been described, and factors that contribute to pharmacological variation in responses can be divided into extrinsic factors like food, cultural practices, and intrinsic factors that include genetics, gender, ancestry, age, body size, renal and hepatic function (Table 1) (1). Amongst these factors, genetic variation has been a prominent contributor to observed phenotypic differences. Since the complete sequencing map of human DNA has become available through the human genome project, insights on human population genetic variation has been greatly enhanced as more coverage of the genome provides more thorough resolution of the genetic structure of populations. The human genome, with 3 billion base pairs encoding more than 30,000 proteins, carries variation of the genetic code called polymorphisms, which contributes largely to phenotypic traits amongst humans. Single nucleotide base pair substitutions (called single nucleotide polymorphisms or SNPs) occur at an average frequency of 1 every 1,000 base pairs. Despite these variations, Homo sapiens are 99.6–99.8% genetically similar to each other. With 0.2% to 0.4% variation, approximately 10 million SNPs exist in the human genome. Much of this understanding of population genetic structure arises from genetic data of autosomes, X-chromosomes and mitochondrial DNA, and reveal that humans who live in proximity to each other geographically have more genetic similarity than those that are separated by distance. Studies of genetic distance can cluster populations into groups, which coincide with self-identified race or continental ancestry. It is clear that between-population genetic variation is less than within-population variation, meaning that most of the SNPs that are present in the human genome can be found within each population. However, SNPs that can distinguish between populations are less frequent. Population-based genetic variations can arise from random mating, genetic drift, distribution and migration—which lead to changes in allelic frequencies. Using a classic measure for analysis of genetic variance within populations relative to between populations—the fixation index (FST), estimates of both “within” and “between” main geographic regions indicate that about 11–23% of variation is due to differences between populations (2-4). Taken together with the 10 million SNPs, this gives still a significant potential for genetic variation between groups of populations, which may bring into effect differences in drug responses between population groups defined by race or ethnicity. Race is defined by the biological characteristics of a person including skin or hair colour and anthropometry, while ethnicity typically refers to identity with cultural, nationality and language.

Full table
How does genetic diversity affect drug pharmacology?
Pharmacokinetics of an administered drug involve processes of absorption, distribution, metabolism and excretion, the processes which determine the concentrations of drug that reach a target tissue and exert its activity through receptor binding. Inter-individual pharmacokinetic variability is usually fairly significant, leading to a continuum of drug pharmacodynamics or responses. Much of this pharmacokinetic variability results from the action of enzymes that metabolise drugs, called drug metabolising enzymes (DMEs). Drug metabolism is divided into two phases: phase 1—reactions that render drugs more reactive or polar, and phase 2—enzymes that conjugate a chemical group to render the drug more water soluble. These reactions can either reduce the pharmacological activity, or increase pharmacological activity of drugs (in the case of pro-drugs). In the circumstance where two populations carrying different frequencies of an allelic variant (with reduced enzymatic activity) of a DME are treated with a standard dose of an oncology drug that undergoes predominant metabolism via this pathway, more frequent on-target adverse events may be seen in the population carrying a higher frequency of the defective allele, due to higher concentration exposure to the active drug. This would be reversed for drugs that are pro-drugs that are activated by the DME to form pharmacologically active derivatives. These allelic variants may be inherited in the homozygous or heterozygous form, resulting in phenotypes called slow and intermediate metabolisers, respectively.
Evidence for population variability of these DME genes derives from next generation sequencing of large number of individuals from major populations, for example in the cytochrome p450 genes, where substantial interethnic variability in frequency of haplotypes that result in altered metabolic phenotypes exist (Table 2) (5,6). Collectively these analyses show that there is justification of using ancestral information in guiding clinical decision-making in the absence of specified genotyping data.
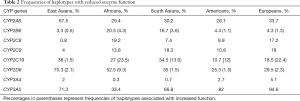
Full table
In oncology, there are many examples of this situation, one of which concerns 6 mercaptopurine (6MP), which is an antineoplastic drug used for acute lymphoblastic leukaemia. 6MP is a prodrug that is activated intracellularly to form active metabolites thioguanine nucleotides, which are then incorporated into DNA and account for its cytotoxicity. 6MP is inactivated by two metabolic pathways, the thiopurine methyltransferase (TPMT) pathway or the xanthine oxidase pathway. The TPMT gene is polymorphic, with 3 populations of patients, those homozygous (0.3% population), heterozygous (~10% population) for non-functional TPMT alleles; homozygous patients develop severe myelosuppression from standard doses of 6MP. There are several common non-functional TPMT alleles: TPMT*2, TPMT*3A, TPMT*3B and TPMT*3C. The pre-emptive genotyping of TPMT to guide dosing of 6MP has increased the safety of 6MP clinically. Whilst the TPMT*3A allele is more common in the Caucasian population (~5%), only the TPMT*3C variant is present in low frequency in East Asians (~1%) (7,8). However, it has been recognised that patients from East Asian populations are more susceptible to 6MP treatment compared to Caucasians, leading the possibility of additional pharmacogenetic influences on 6MP treatment. Subsequent genome wide association studies (GWAS) showed that this could be explained by pharmacogenetics of another previously unrecognised gene, NUDT15, which encodes an enzyme that was found to be able to deactivate TGTP to TGMP. NUDT15 is highly polymorphic, with 4 coding variants leading to defective activity, and specifically, the presence of an pArg139Cys variant could account for the observed East Asian susceptibility to 6MP compared to Caucasians. In fact, the frequency of the low or intermediate diplotypes of NUDT15 occurred in 22.6% of East Asians, making it very pertinent to genotype for this variant prior to 6MP use. Therefore, using observed ethnic differences in drug phenotypes can lead to novel pharmacogenetics discoveries, rendering safety to the use of these drugs in the general populations.
Irinotecan, a topoisomerase 1 inhibitor that is standard of care in colorectal carcinoma, undergoes activation to its active metabolite, SN38, which is predominantly inactivated via glucuronidation through the same enzyme that glucuronidates bilirubin, UGT1A1. This enzyme is polymorphic, and reduced activity is found in patients with Gilbert’s syndrome, where hyperbilirubinemia is observed. It has been shown that patients who harbour the variant UGT1A1*28 allele where 7 tandem TA repeats are found in the promoter region of the gene compared to 6 repeats are more prone to life threatening neutropenia from standard doses of irinotecan. This polymorphism demonstrates racial variation, with highest frequency in African Americans, followed by Caucasians, and is relatively less common in East Asian populations. On the other hand, other allelic variants (UGT1A1*6 and UGT1A1*27) that impair UGT1A1 function are more common in East Asians than Caucasians (9-12).
Some oncology drugs exhibit clear population variability with regards to adverse effects, but the exact reasons for this are largely still unclear. The example is 5-fluorouracil (5FU), where differences in clinical tolerability have been described according to geographical regions of the world treated with this agent or its prodrug, capecitabine, for colorectal cancer. Ethnic differences also exist in 5FU tolerability with significantly higher rates of haematological toxicity seen in African American population as compared to Caucasians. This may be related to lower levels of dihydropyrimidine dehydrogenase (DPD) seen in peripheral mononuclear cells in African Americans when compared to Caucasians (13). In comparison, 5FU and its oral pro-drug capecitabine are more tolerable for East Asian patients, possibly relating to polymorphisms in the gene encoding thymidylate synthase (TYMS). Variations in tandem repeat sequence in the promoter enhancer region are seen, commonly in duplet (2R) or triplet (3R) form. Increased TYMS expression is associated with 3R expression and the 3R/3R genotype is common in Chinese individuals compared to their Caucasian counterparts. Patients with 2R/2R genotype are at increased risk of treatment associated toxicities. In a study of 1,864 patients, severe grade 3 to 4 gastrointestinal toxicity presenting as mucositis and diarrhoea was 3.62 times more frequent in United States (US) patients compared to East Asian patients. Interestingly, the incidence of grade 3 to 4 neutropenia was found to be the same in both groups of patients. It is known that 5FU is inactivated by the rate-limiting enzyme DPD, an enzyme that is polymorphic and there is a discrepancy between population frequencies of genetic variants (<5%) and phenotype of functional DPD deficiency (up to 20%). Nonetheless, individuals genotyped with DPYD2A (IVS14+1G>A), DPD*13 (c.1679T>G) and c.2846A>T account for cases of very severe toxicities to 5FU treatment. About 10–40% patients develop severe and potentially life-threatening toxicity early during 5FU treatment (14). Variability in population frequencies of several of these variant alleles have been described; however, their contributions to observed differences in clinical toxicities is unclear.
Another oral prodrug of 5FU, TS-One, is a pharmacological combination of tegafur, a prodrug of 5FU, gimeracil, a potent DPD inhibitor (to enhance bioavailability of 5FU after conversion of tegafur to 5FU), and oteracil, a poorly absorbable inhibitor of pyrimidine phosphoribosyl transferase (PPRT), the activating enzyme for 5FU. In a small comparison study of TS-One in Singaporean Asians and US Caucasians, gastrointestinal toxicities were found to be higher in frequency in Caucasians. Interestingly, as tegafur is converted to 5FU in the liver through the polymorphic enzyme CYP2A6, it was shown that this conversion was correlated with nicotine metabolism, and that polymorphism of CYP2A6 will impact on the activation of tegafur. However, the overall 5FU plasma concentrations in both study populations were similar (15). Therefore, the source of variability of gastrointestinal (GI) toxicities between populations treated with 5FU is currently unclear.
As with most pharmaceutical agents, oncology drugs that undergo hepatic metabolism utilise the cytochrome p450 family of isoenzymes. Many of these subfamilies of enzymes are polymorphic, including CYP2D6, CYP2A6, CYP2C19, and CYP2C9. A scan of the frequency distribution of these polymorphisms shows quite marked differences across racial groups (Table 2). Consistent with genome maps, African populations carry the most degrees of variability. These will have profound influence on the pharmacokinetics and pharmacodynamics of drugs predominantly metabolised through the relevant pathways.
Another way which genetic variability can affect drug responses is the sensitivity of the target receptor to the drug. These receptors are subject to mutations that affect their function and binding affinity to drugs. Therefore, variation in frequencies of these drug receptor-related mutations may result in marked differences in response to standard doses of drug treatment. A classic example is in warfarin where the target of warfarin, the VKORC1 gene haplotypes H1 and H2 result in reduced expression of the enzyme and therefore render patients with these haplotypes sensitive to warfarin, and therefore higher propensity to over-anticoagulation (16).
In oncology, the somatic EGFR mutations at the TK domain render tumours carrying these mutations more sensitive to EGFR tyrosine kinase inhibitors (TKIs). It is notable that these mutations, though not germline, are 2–3 fold more frequent in non-small cell lung cancer (NSCLC) in East Asians (35%) compared with Caucasians (15%), a difference persists in patients of Asian ancestry living in North America (17). The reason for this discrepancy is unknown. An element of ethno-geographical variability is also seen in KRAS mutational status in colorectal cancer, with lower levels seen in Asian patients compared to European and North American rates (18). On the other hand, driver events other than EGFR mutations like ALK/ROS1/RET gene rearrangements in non-squamous lung cancer do not appear to show ethno-geographic variability.
SNPs are not the only genetic events that can affect activity of DMEs. The most common mutations that occur in the genome result in alterations in the copy numbers of the gene [copy number variation (CNV)], and therefore affect the extent of gene expression; more copy numbers of the genes lead to higher expression and therefore function. It could arise by deletion of a part of the DNA resulting loss of a gene, or duplication of a gene. In terms of base coverage, CNV forms the majority of genetic diversity in the human genome. Geographical variation of these CNV have been described through comprehensive chromosomal comparative genomic hybridisation (cCGH) combined with next generation sequencing. Environmental pressure and selection could to a certain extent contribute to these geographical variations. For example, CNV of the amylin gene that catabolises carbohydrates have been found to be higher in populations dependent on starch compared to those whose diet is mainly meat (19). Response to drug treatment may be affected by increased or loss of copies of DME genes. The overall effect is an allelic gain or loss leading to alteration in the expression of the particular metabolic enzyme, thus increasing or reducing the function of the enzyme. For drugs that are inactivated, higher doses may be necessary in such individuals to achieve the same pharmacodynamic effect, whereas for prodrugs, lower doses will achieve the same effect as standard doses in wild type individuals. In a phase 2 isoenzyme UDP-glucuronosyltransferase (UGT) 2B17, a deletion of a 150 kb DNA including the gene results in a polymorphism called UGT2B17*2 and demonstrates very marked geographical variation—it is very common in the East Asian populations where homozygous UGT2B17*2/*2 reaches 67% of the Korean population and almost 80% of the Japanese population, compared with 9–15% of the Caucasian population (20). UGT2B17 is involved in phase 2 metabolism of glucuronidation of xenobiotics like coumarins, anthroquinones and flavonoids. Several oncology agents like the histone deacetylase inhibitor vorinostat and the irreversible steroid-based aromatase inhibitor exemestane are metabolised by UGT2B17. It is the major enzyme responsible for glucuronidation and inactivation of the active metabolite of exemestane and presence of UGT2B17*2/*2 null genotype has a significant effect on exemestane pharmacokinetics (21).
Transporter expression
Members of the ATP-binding cassette (ABC) transporter family are involved in transport and excretion of chemotherapeutic agents and multiple polymorphisms exist known to affect function. SNP C421A leads to reduced expression and substrate specific binding and shows variation depending on ancestry with frequencies of 27–35% in Asians, 9–14% in Caucasians, and 1–5% in Africans. Presence of heterozygosity for SNP C421A is shown to increase accumulation of substrates including gefitinib in affected individuals and is associated with increased rates of related toxicities (22).
Impact of human leukocyte antigen (HLA)
Immunotherapy has changed the landscape of treatment of many tumour groups including NSCLC, melanoma and renal cell cancer (RCC) among others, however defining the population most likely to benefit from such therapy is challenging. Programmed death-ligand 1 (PD-L1) tumour proportion score (TPS) is approved for use in NSCLC to direct use of the immune checkpoint inhibitor (ICI) pembrolizumab. However, there are a large proportion of patients who exhibit high PD-L1 TPS who fail to respond to pembrolizumab monotherapy and the search for improved biomarkers is ongoing.
The HLA class 1 and 2 loci are highly polymorphic, with great variation in frequencies of HLA genotypes according to ethnic population and geography (23). With nucleotide diversity at the peptide binding regions of HLA genes, effect on antigen presentation would likely affect immune responses. There is significant correlation between the global patterns of HLA nucleotide diversity among populations with ethnicity and geography. In patients with melanoma treated with ICIs better survival is seen in patients who have heterozygosity for at least one locus in the HLA class 1 genes, suggesting that diversity at this locus influences tumour neoantigen presentation and immune activation. Specific HLA alleles are also noted to be associated with improved outcomes to ICI therapy, for example HLA B44 superfamily was associated with improved survival in melanoma patients treated with ICI (24).
Toxicities as a result of ICI therapy are seen to vary among populations. Analysis of data from “PACIFIC” study, a phase III study of Durvalumab in stage III unresectable NSCLC, shows increased rates of pneumonitis in Japanese patients (25). An apparently high rate of pneumonitis was also seen in a second line study of nivolumab for advanced non-squamous lung carcinoma in Japanese patients when compared to rates in comparison studies in Caucasian populations (26,27). The underlying cause for this variability is not clear.
Other sources of ethnic variability
Further underscoring the fact that it may be difficult to elucidate the sources of variability despite clear observations of ethnic variability of response and outcome to drug treatment is a retrospective analysis of survival outcomes of children with acute lymphoblastic leukaemia in the US treated with the Children’s Cancer Group protocols (28). Confounding factors including clinical features, disease biology, era of treatment and socioeconomic status were accounted for. The analysis showed an association between ethnicity and long-term survival outcomes with Asian children having the best 5-year event-free survival rates, while survival of black children fares comparatively worst. There is a need to understand these differences, whether they are down to unmeasured confounding factors such as treatment compliance versus underlying pharmacogenetics. In another study of a commonly used regimen of three-weekly administered docetaxel and carboplatin for metastatic NSCLC, the combination was found to be more myelosuppressive in an East Asian compared to a Caucasian cohort; interestingly, efficacy was better in the Asian patients (29). Reasons for these differences are not well understood currently.
Disease etiology and characteristics can lead to differences in drug treatment outcome in different populations of patients. For example, the multi-kinase inhibitor sorafenib is approved globally for management of hepatocellular carcinoma (HCC); in the Western populations, sorafenib improved the overall survival of these patients, with median overall survival of treated patients reaching 10.7 months. with a hazard ratio of 0.69. In Asian patients, the median overall survival of patients with HCC was shorter at 6.5 months. This is due to the higher incidence of hepatitis B related HCC in Asian populations, with this group of patients having poorer prognosis. In fact, sorafenib treatment had a similar hazard ratio of improvement in both populations, 0.69 in the Western study and 0.68 in the Asian study.
Body size characteristics can vary between ethnic groups, and would potentially affect drugs where exposure and effects are related to body size measures (30). For example, in monoclonal antibodies dosed by body weight, and to apply flat dosing based on average body weight has to take into account the differences in population body weights.
Issues with ethnicity and race as a biomarker for drug response
The real issues of population grouping by race and ethnicity are several. Firstly, scientists have difficulty in ascribing race and ethnicity in consistent scientific methodology. While most phenotypic traits can be measured, race and ethnicity are difficult to define as they are social constructs. The situation becomes more complex with population admixture, leading to genes sharing between racial groups, rendering it difficult to ascribe genotypes based on racial intuition compared to genotyping. An example is a study of the genotypes of 2 pioneers of genetics, James Watson and Craig Venter. Watson, ostensibly Caucasian, had two polymorphisms in CYP2D6 that are supposedly found more frequently in East Asians (31).
Indeed, to avoid ethical issues of political segregation, racial disparity in treatment access, scientists have questioned the use of race or ethnicity as factors for biomedical investigation, and considered race to be a poor proxy for genetic variation. For this reason, it has been considered more appropriate to label a person according to his or her ancestry. In the US, race classification commonly follows five major groups: African American, white, Asian, native Hawaiian or other Pacific Islanders. However, this does not take into account variability within each group: for example, it is well established that South Asians and West Asians are genetically more similar to European and Western populations than East Asians. Indeed, a situation where legal action is posed by populations where drugs are perceived to be less efficacious due to frequency of a variant allele is regrettable (32). A significant proportion of FDA approved medications involve metabolism by genes that are known to be have population differences in polymorphisms.
Addressing population differences in drug responses
Despite the issues with using racial, ethnic or even ancestral data to stratify patients for clinical drug treatment decisions, to ignore race, ethnicity or ancestry in study of variation of drug responses would be an opportunity lost in pharmacogenetic discovery, where genetic determinants of this variability could be used to predict tolerability, reduce toxicity and enhance efficacy. While in current clinical practice physicians do adjust patients’ drug dosing according to their ancestry in the absence of genotyping, studying population differences in drug responses and resolving these based on genetic variation leads to the potential for personalised precision therapeutics to optimise outcomes and reduce toxicities. To make this reality, pre-emptive pharmacogenomics will need to be applied in a clinically palatable way for physicians.
Ethno-bridging studies have been a proposed way to ensure safety and tolerability of drug dosing and treatment based on data obtained from other study populations. These studies utilise data from across ethnic cohorts of patients. There have been country specific regulations by healthcare authorities with regards to application of data obtained from foreign patient population. To provide a framework to guide the factors to consider when determining the need for bridging studies when foreign clinical trial data is used for drug registration, the FDA issued the E5 document in 1998. This guidance document aimed to help with decision making on regulatory and further development when extrapolating clinical data of medications between ethnically diverse regions, recognising that it is important to reduce the need for duplication of clinical trial efforts where possible, and expedite access to new medicines (33).
Drugs that are deemed sensitive to ethnic differences include those listed in Table 3. These are often characteristics in cancer treatment drugs, and should be considered when determining the need for bridging studies.

Full table
Pharmaceutical companies are encouraged currently to include global populations into their multi-national clinical trials as early as possible, to collect data from geographically separated populations and detect differences to drug response early. Data from patients of an ethnicity to support registration of a drug in the country populated by that particular ethnic group can help accelerate drug development. Study design should take into account pharmacokinetic, pharmacodynamic and pharmacogenetics in relation to ethnic groups in drug development.
Summary
Clearly population variation in drug responses in oncology have been described, and should be considered in drug development phases. Ways to address this may be to include sufficient diverse population representation during drug development, include predetermined statistical analysis for population comparisons. Including pharmacokinetics and genotype can help greatly to understand the sources of this variability, with the ultimate objective of developing genetic biomarkers for individualised therapy.
Acknowledgments
None.
Footnote
Conflicts of Interest: The authors have no conflicts of interest to declare.
References
- Schuck RN, Florian J, Charlab R, et al. Trial geography, pharmacogenetics, and global drug development. Clin Pharmacol Ther 2015;97:218-20. [Crossref] [PubMed]
- Tishkoff SA, Verrelli BC. Patterns of human genetic diversity: implications for human evolutionary history and disease. Annu Rev Genomics Hum Genet 2003;4:293-340. [Crossref] [PubMed]
- Kidd KK, Pakstis AJ, Speed WC, et al. Understanding human DNA sequence variation. J Hered 2004;95:406-20. [Crossref] [PubMed]
- Tishkoff SA, Williams SM. Genetic analysis of African populations: human evolution and complex disease. Nat Rev Genet 2002;3:611-21. [Crossref] [PubMed]
- Zhou Y, Ingelman-Sundberg M, Lauschke VM. Worldwide Distribution of Cytochrome P450 Alleles: A Meta-analysis of Population-scale Sequencing Projects. Clin Pharmacol Ther 2017;102:688-700. [Crossref] [PubMed]
- Ramos E, Doumatey A, Elkahloun AG, et al. Pharmacogenomics, ancestry and clinical decision making for global populations. Pharmacogenomics J 2014;14:217-22. [Crossref] [PubMed]
- Collie-Duguid ES, Pritchard SC, Powrie RH, et al. The frequency and distribution of thiopurine methyltransferase alleles in Caucasian and Asian populations. Pharmacogenetics 1999;9:37-42. [Crossref] [PubMed]
- Chang JG, Lee LS, Chen CM, et al. Molecular analysis of thiopurine S-methyltransferase alleles in South-east Asian populations. Pharmacogenetics 2002;12:191-5. [Crossref] [PubMed]
- Ando Y, Saka H, Ando M, et al. Polymorphisms of UDP-glucuronosyltransferase gene and irinotecan toxicity: a pharmacogenetic analysis. Cancer Res 2000;60:6921-6. [PubMed]
- Bosma PJ, Chowdhury JR, Bakker C, et al. The genetic basis of the reduced expression of bilirubin UDP-glucuronosyltransferase 1 in Gilbert's syndrome. N Engl J Med 1995;333:1171-5. [Crossref] [PubMed]
- Araki K, Fujita K, Ando Y, et al. Pharmacogenetic impact of polymorphisms in the coding region of the UGT1A1 gene on SN-38 glucuronidation in Japanese patients with cancer. Cancer Sci 2006;97:1255-9. [Crossref] [PubMed]
- Shimoyama S. Pharmacogenetics of irinotecan: An ethnicity-based prediction of irinotecan adverse events. World J Gastrointest Surg 2010;2:14-21. [Crossref] [PubMed]
- Mattison LK, Fourie J, Desmond RA, et al. Increased prevalence of dihydropyrimidine dehydrogenase deficiency in African-Americans compared with Caucasians. Clin Cancer Res 2006;12:5491-5. [Crossref] [PubMed]
- Amstutz U, Farese S, Aebi S, et al. Dihydropyrimidine dehydrogenase gene variation and severe 5-fluorouracil toxicity: a haplotype assessment. Pharmacogenomics 2009;10:931-44. [Crossref] [PubMed]
- Chuah B, Goh BC, Lee SC, et al. Comparison of the pharmacokinetics and pharmacodynamics of S-1 between Caucasian and East Asian patients. Cancer Sci 2011;102:478-83. [Crossref] [PubMed]
- Rieder MJ, Reiner AP, Gage BF, et al. Effect of VKORC1 haplotypes on transcriptional regulation and warfarin dose. N Engl J Med 2005;352:2285-93. [Crossref] [PubMed]
- Soh J, Toyooka S, Matsuo K, et al. Ethnicity affects EGFR and KRAS gene alterations of lung adenocarcinoma. Oncol Lett 2015;10:1775-82. [Crossref] [PubMed]
- Yoon HH, Shi Q, Alberts SR, et al. Racial Differences in BRAF/KRAS Mutation Rates and Survival in Stage III Colon Cancer Patients. J Natl Cancer Inst 2015. [Crossref] [PubMed]
- Perry GH, Dominy NJ, Claw KG, et al. Diet and the evolution of human amylase gene copy number variation. Nat Genet 2007;39:1256-60. [Crossref] [PubMed]
- Jakobsson J, Ekström L, Inotsume N, et al. Large differences in testosterone excretion in Korean and Swedish men are strongly associated with a UDP-glucuronosyl transferase 2B17 polymorphism. J Clin Endocrinol Metab 2006;91:687-93. [Crossref] [PubMed]
- Walsh RJ, Lee SC, Seng KY, et al. Prospective study of UDP-glucuronosyltransferase (UGT) 2B17 genotype and exemestane (Exe) pharmacokinetics (PK) and pharmacodynamics (PD) in Asian, hormone receptor (HR) positive, metastatic breast cancer (MBC) patients. J Clin Oncol 2017;35:abstr 1056.
- Ahmed S, Zhou Z, Zhou J, et al. Pharmacogenomics of Drug Metabolizing Enzymes and Transporters: Relevance to Precision Medicine. Genomics Proteomics Bioinformatics 2016;14:298-313. [Crossref] [PubMed]
- Buhler S, Sanchez-Mazas A. HLA DNA sequence variation among human populations: molecular signatures of demographic and selective events. PLoS One 2011;6:e14643. [Crossref] [PubMed]
- Chowell D, Morris LGT, Grigg CM, et al. Patient HLA class I genotype influences cancer response to checkpoint blockade immunotherapy. Science 2018;359:582-7. [Crossref] [PubMed]
- Antonia SJ, Villegas A, Daniel D, et al. Durvalumab after Chemoradiotherapy in Stage III Non-Small-Cell Lung Cancer. N Engl J Med 2017;377:1919-29. [Crossref] [PubMed]
- Nishio M, Hida T, Atagi S, et al. Multicentre phase II study of nivolumab in Japanese patients with advanced or recurrent non-squamous non-small cell lung cancer. ESMO Open 2017;1:e000108. [PubMed]
- Borghaei H, Paz-Ares L, Horn L, et al. Nivolumab versus Docetaxel in Advanced Nonsquamous Non-Small-Cell Lung Cancer. N Engl J Med 2015;373:1627-39. [Crossref] [PubMed]
- Bhatia S, Sather HN, Heerema NA, et al. Racial and ethnic differences in survival of children with acute lymphoblastic leukemia. Blood 2002;100:1957-64. [Crossref] [PubMed]
- Millward MJ, Boyer MJ, Lehnert M, et al. Docetaxel and carboplatin is an active regimen in advanced non-small-cell lung cancer: a phase II study in Caucasian and Asian patients. Ann Oncol 2003;14:449-54. [Crossref] [PubMed]
- Walpole SC, Prieto-Merino D, Edwards P, et al. The weight of nations: an estimation of adult human biomass. BMC Public Health 2012;12:439. [Crossref] [PubMed]
- Ng PC, Zhao Q, Levy S, et al. Individual genomes instead of race for personalized medicine. Clin Pharmacol Ther 2008;84:306-9. [Crossref] [PubMed]
- Wu AH, White MJ, Oh S, et al. The Hawaii clopidogrel lawsuit: the possible effect on clinical laboratory testing. Per Med 2015;12:179-81. [Crossref] [PubMed]
- The Food and Drug Administration. E5 ethnic factors in the acceptability of foreign clinical data. 2004. Available online: https://www.fda.gov/regulatory-information/search-fda-guidance-documents/e5-ethnic-factors-acceptability-foreign-clinical-data


