
The impact of somatic SMAD4 mutations in colorectal liver metastases
Introduction
Colorectal cancer (CRC) is the second leading cause of cancer-related death and the third most frequently diagnosed cancer in the United States (1). While the prognosis for CRC varies widely, approximately 20% of patients with CRC have metastases at the time of diagnosis. For patients who develop colorectal liver metastases (CRLM), multimodality therapy consisting of contemporary chemotherapy and metastasectomy can lead to 5-year overall survival (OS) rates that exceed 50% (2,3). Nevertheless, more than half of these patients will develop recurrent disease within 2 years from surgery (4). Therefore, the accurate risk stratification of patients for recurrence, via the identification of relevant prognostic factors, may lead to improved outcomes through individualized treatment or specific surveillance strategies.
In recent years, significant advances have led to the discovery and characterization of specific genes and molecular pathways involved in carcinogenesis, disease progression, and mechanisms of treatment resistance in CRC biology. While these advances have led to the ability to provide improved prognostic information, novel biomarkers based on genomic profiling are required to further differentiate among the various subgroups of patients with CRC. More importantly, these genome-oriented biomarkers for CRC would facilitate the implementation of more personalized treatment, potentially leading to better prognosis and improved survival.
SMAD4 in CRC progression
Alterations of the transforming growth factor (TGF)-β signaling cascade play an essential role in carcinogenesis and disease progression of CRC given its critical involvement in cell proliferation, differentiation, apoptosis, and extracellular matrix production (5,6). While activation of TGFβ may be associated with tumor suppression in early stages (7), it is hypothesized to promote angiogenesis, epithelial-to-mesenchymal transition, and tumor metastasis in later stages of CRC development (8,9).
Drosophila protein, mothers against decapentaplegic homolog 4 (SMAD4) is an essential mediator in the TGFβ signaling pathway, which is located on chromosome 18q21 (5). Following the linking of TGFβ ligands to TGFβ transmembrane protein kinase receptors 1 and 2, SMAD2 and SMAD3 proteins are activated via phosphorylation which subsequently allows them to link to SMAD4 (10,11). The activated SMAD4 complex then relocates to the nucleus where it regulates TGFβ-related gene transcription (6,12-15). Among the components of this protein cascade, SMAD4 is an essential intermediator, exhibiting a critical role as a common downstream regulator and tumor suppressor gene (16,17) (Figure 1).
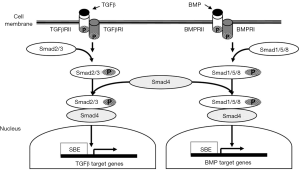
SMAD4 mutations that lead to decreased SMAD4 protein expression have been reported to occur in approximately 20% of patients with CRC (18-20). The loss of SMAD4 expression has been implicated both in metastasis and in poor response to chemotherapy for patients with CRC (21-24). In fact, recent studies have demonstrated that decreased SMAD4 expression is independently associated with worse recurrence-free survival (RFS) and OS among patients with CRC, especially among those with advanced stages of disease (22,25-27). In contrast, greater levels of SMAD4 expression are associated with improved disease-free survival (DFS) and OS (26). It is posited that SMAD4 inactivation leads to unregulated TGFβ-induced growth (23) which may contribute to worse prognosis in CRC (28).
Recent work has also highlighted that SMAD4 downregulation may occur in up to 60% of patients with metastatic CRC, which is significantly higher than the incidence of SMAD4 mutations (29). Interestingly, there is increasing evidence that SMAD4 expression is also regulated by several micro-RNAs (30). Specifically, previous studies using animal models have noted that SMAD4 is influenced by miR-130a, miR-20a, miR-224, miR-34a and miR-19b (31-35). While the exact mechanism of action and role of these micro-RNAs in regulating SMAD4 in metastatic CRC progression is not understood, there is evidence that micro-RNAs contribute to chemotherapy-resistance. For example, several studies have indicated that the upregulation of miR-20a, miR-224 and miR-19b is associated with decreased response to the commonly used chemotherapy agent 5-fluorouracil (5-FU) (36-38). Similarly, these same micro-RNAs reduce the sensitivity of CRC cells to oxaliplatin, another commonly employed chemotherapeutic in CRC (35,36,39). Therefore, ongoing research on the regulatory mechanisms of miRNAs to the downregulation of SMAD4 could have important and relevant therapeutic implications to the management of patients with metastatic CRC.
It is equally important to note that somatic mutations, concurrent with SMAD4, may have an additive effect on the prognosis of patients with CRC. For example, concomitant SMAD4 and PTEN mutations have been identified in a subgroup of patients with CRC who have more aggressive disease. This gene association is believed to yield significantly worse outcomes compared with patients with CRC who have only one of these mutations (40). Interestingly, there is evidence that micro-RNAs miR-130a (41), miR-20a (42) and miR-19b (43) regulate PTEN in addition to SMAD4. Nevertheless, the precise role of the specific markers that simultaneously decrease the expression of SMAD4 and PTEN and contribute to the development of more aggressive CRC or how exactly they yield resistance to 5-FU and oxaliplatin treatments have not been explicitly elucidated (35,36,38).
Besides the association with poor prognosis in CRC, SMAD4 mutations have been also found to be associated with colonic tumor location, female sex and mucinous histology type (44,45). In more recent studies, SMAD4 mutation was found to be associated with high- versus low-grade mucinous adenocarcinomas, advanced stage of disease and aggressive phenotypes of CRC (22,46-48). However, as previously discussed, the worse outcomes observed in patients with CRC may be in part confounded by the relative resistance to 5-FU (26,49).
SMAD4 mutations in CRLM
The incidence of somatic SMAD4 mutations in patients with isolated CRLM is approximately 15%. Recently published data on surgical outcomes for patients with CRLM have also highlighted the clinical relevance of SMAD4 mutations among patients undergoing liver resection (50). Mizuno et al. retrospectively evaluated the outcomes of patients with known SMAD4 gene mutation status following hepatectomy for CRLM. SMAD4 mutations were found to be independently associated with worse OS following liver resection, independent of RAS mutation status. Furthermore, the negative prognostic impact of SMAD4 gene mutation status was also confirmed in a validation cohort of patients who only received systemic chemotherapy for metastatic CRC (50). The results of this study highlight the utility of SMAD4 mutation status in the surgical decision-making for patients scheduled to undergo surgical resection for CRLM, especially patients with initially unresectable disease or those who are scheduled to undergo a two-stage hepatectomy.
While the impact of SMAD4 mutation status on recurrence rates or DFS in patients with CRLM undergoing hepatectomy has not been yet clearly established, it is notable that patients with SMAD4 mutations were less likely to undergo repeat hepatectomy for recurrent disease after initial metastasectomy (50). Although the survival differences after recurrence between patients with SMAD4 gene mutations versus SMAD4 wild type tumor genotype may represent differences in ability to undergo repeat hepatectomy, the mechanisms driving this difference is unknown and warrants additional investigation (51,52). A plausible explanation is that SMAD4 mutant recurrences are more likely to occur in an unresectable fashion due to their more aggressive tumor biology, or alternatively, they may become unresectable because of tumor progression during neoadjuvant chemotherapy.
It is also known that patients with SMAD4-mutant CRLM also have a higher incidence of somatic RAS mutations. This is presumably related to differences in the specific signaling pathways that each protein mediates: while SMAD4 regulates the TGFβ signaling pathway (5), RAS plays an important role in the mitogen-activated protein kinase signaling pathway (53). Nevertheless, the synergistic action between SMAD4 and RAS should be further investigated since both gene mutations are important independent predictors of poor OS (50). Additional research into the mechanistic and prognostic importance of other somatic mutations in CRLM, such as APC, PIK3CA, BRAF and TP53, will only enhance our understanding regarding the complex molecular pathways involved in CRC development, progression, prognosis, and response to treatment (54,55). In fact, emerging evidence suggests that mutation status of RAS, TP53, and SMAD4 provides superior prognostic information following resection of CRLM compared to any single or double somatic mutation alone (56).
Future research
While the prognostic importance of somatic mutations in CRC and more specifically in CRLM continues to be highlighted, future research is necessary to address several unanswered questions. The distinct molecular mechanisms underlying the complex SMAD4 regulatory network, including the specific mechanisms by which micro-RNAs downregulate SMAD4 expression and leads to the disruption of important TGFβ signaling pathways, need to be better understood. A detailed elucidation of these mechanisms will aid the translation of foundational molecular concepts into the establishment of future novel, targeted, therapeutic advances for patients with CRC. The clinical significance of SMAD4 and its influence on the use of diverse and individualized perioperative therapies for CRC need to be validated by future investigations. An important question to answer is whether the SMAD4 gene can be specifically targeted as a novel therapeutic agent for patients with CRC. Given the evidence that concomitant mutations in RAS, TP53, APC and PIK3CA are associated with worse OS following hepatectomy for CRLM, the precise cooperative mechanisms of SMAD4 with other genes of influence requires further examination. Future research is also warranted regarding the gene’s impact on the recurrence rates for patients with CRLM undergoing hepatectomy.
Conclusions
In summary, SMAD4 expression mediates an important role in the development and progression of CRC. Somatic mutations of SMAD4 are associated with more aggressive tumor biology, poor response to chemotherapy, metastases and unfavorable OS among patients with resectable and unresectable CRC. Additionally, there is evidence that SMAD4 mutations are significantly associated with worse OS, irrespective of RAS mutation status or other clinicopathological factors, in patients undergoing metastasectomy for CRLM. Given the relative frequency with which SMAD4 mutations occur among patients with CRC, routine SMAD4 testing may be appropriate. In the contemporary era of personalized treatment for CRC, further research on whether SMAD4 represents a targetable mutation could have important implications for guiding clinical-decision making.
Acknowledgments
None.
Footnote
Conflicts of Interest: The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
References
- Siegel RL, Miller KD, Jemal A. Cancer statistics, 2017. CA Cancer J Clin 2017;67:7-30. [Crossref] [PubMed]
- Abdalla EK, Vauthey JN, Ellis LM, et al. Recurrence and outcomes following hepatic resection, radiofrequency ablation, and combined resection/ablation for colorectal liver metastases. Ann Surg 2004;239:818-25. [Crossref] [PubMed]
- Brouquet A, Abdalla EK, Kopetz S, et al. High survival rate after two-stage resection of advanced colorectal liver metastases: response-based selection and complete resection define outcome. J Clin Oncol 2011;29:1083-90. [Crossref] [PubMed]
- de Jong MC, Pulitano C, Ribero D, et al. Rates and patterns of recurrence following curative intent surgery for colorectal liver metastasis: an international multi-institutional analysis of 1669 patients. Ann Surg 2009;250:440-8. [PubMed]
- Lagna G, Hata A, Hemmati-Brivanlou A, et al. Partnership between DPC4 and SMAD proteins in TGF-beta signalling pathways. Nature 1996;383:832-6. [Crossref] [PubMed]
- Malkoski SP, Wang XJ. Two sides of the story? SMAD4 loss in pancreatic cancer versus head-and-neck cancer. FEBS Lett 2012;586:1984-92. [Crossref] [PubMed]
- Ikushima H, Miyazono K. TGFbeta signalling: a complex web in cancer progression. Nat Rev Cancer 2010;10:415-24. [Crossref] [PubMed]
- Pickup M, Novitskiy S, Moses HL. The roles of TGFbeta in the tumour microenvironment. Nat Rev Cancer 2013;13:788-99. [Crossref] [PubMed]
- Drabsch Y, ten Dijke P. TGF-beta signalling and its role in cancer progression and metastasis. Cancer Metastasis Rev 2012;31:553-68. [Crossref] [PubMed]
- Abdollah S, Macias-Silva M, Tsukazaki T, et al. TbetaRI phosphorylation of Smad2 on Ser465 and Ser467 is required for Smad2-SMAD4 complex formation and signaling. J Biol Chem 1997;272:27678-85. [Crossref] [PubMed]
- Souchelnytskyi S, Tamaki K, Engstrom U, et al. Phosphorylation of Ser465 and Ser467 in the C terminus of Smad2 mediates interaction with SMAD4 and is required for transforming growth factor-beta signaling. J Biol Chem 1997;272:28107-15. [Crossref] [PubMed]
- Shi Y, Hata A, Lo RS, et al. A structural basis for mutational inactivation of the tumour suppressor SMAD4. Nature 1997;388:87-93. [Crossref] [PubMed]
- Chacko BM, Qin B, Correia JJ, et al. The L3 loop and C-terminal phosphorylation jointly define Smad protein trimerization. Nat Struct Biol 2001;8:248-53. [Crossref] [PubMed]
- Chacko BM, Qin BY, Tiwari A, et al. Structural basis of heteromeric smad protein assembly in TGF-beta signaling. Mol Cell 2004;15:813-23. [Crossref] [PubMed]
- Xu Y, Pasche B. TGF-beta signaling alterations and susceptibility to colorectal cancer. Hum Mol Genet 2007;16:R14-20. [Crossref] [PubMed]
- Massaous J, Hata A. TGF-beta signalling through the Smad pathway. Trends Cell Biol 1997;7:187-92. [Crossref] [PubMed]
- Derynck R, Zhang Y, Feng XH. Smads: transcriptional activators of TGF-beta responses. Cell 1998;95:737-40. [Crossref] [PubMed]
- Riggins GJ, Kinzler KW, Vogelstein B, et al. Frequency of Smad gene mutations in human cancers. Cancer Res 1997;57:2578-80. [PubMed]
- Fearon ER, Cho KR, Nigro JM, et al. Identification of a chromosome 18q gene that is altered in colorectal cancers. Science 1990;247:49-56. [Crossref] [PubMed]
- Salovaara R, Roth S, Loukola A, et al. Frequent loss of SMAD4 /DPC4 protein in colorectal cancers. Gut 2002;51:56-9. [Crossref] [PubMed]
- Yamamoto T, Kawada K, Itatani Y, et al. Loss of SMAD4 Promotes Lung Metastasis of Colorectal Cancer by Accumulation of CCR1 Tumor-Associated Neutrophils through CCL15-CCR1 Axis. Clin Cancer Res 2017;23:833-44. [Crossref] [PubMed]
- Miyaki M, Iijima T, Konishi M, et al. Higher frequency of SMAD4 gene mutation in human colorectal cancer with distant metastasis. Oncogene 1999;18:3098-103. [Crossref] [PubMed]
- Papageorgis P, Cheng K, Ozturk S, et al. SMAD4 inactivation promotes malignancy and drug resistance of colon cancer. Cancer Res 2011;71:998-1008. [Crossref] [PubMed]
- Zhang B, Halder SK, Kashikar ND, et al. Antimetastatic role of SMAD4 signaling in colorectal cancer. Gastroenterology 2010;138:969-80.e1. [Crossref] [PubMed]
- Roth AD, Delorenzi M, Tejpar S, et al. Integrated analysis of molecular and clinical prognostic factors in stage II/III colon cancer. J Natl Cancer Inst 2012;104:1635-46. [Crossref] [PubMed]
- Alazzouzi H, Alhopuro P, Salovaara R, et al. SMAD4 as a prognostic marker in colorectal cancer. Clin Cancer Res 2005;11:2606-11. [Crossref] [PubMed]
- Alhopuro P, Alazzouzi H, Sammalkorpi H, et al. SMAD4 levels and response to 5-fluorouracil in colorectal cancer. Clin Cancer Res 2005;11:6311-6. [Crossref] [PubMed]
- Voorneveld PW, Jacobs RJ, Kodach LL, et al. A meta-analysis of SMAD4 immunohistochemistry as a prognostic marker in colorectal cancer. Transl Oncol 2015;8:18-24. [Crossref] [PubMed]
- Losi L, Luppi G, Benhattar J. Assessment of K-ras, SMAD4 and p53 gene alterations in colorectal metastases and their role in the metastatic process. Oncol Rep 2004;12:1221-5. [PubMed]
- Yuan X, Zhang Z, Jiang K, et al. Preliminary study of the role F-Box protein 32 (FBXO32) in colorectal neoplasms through the transforming growth factor beta (TGF-b)/SMAD4 signalling pathway. Med Sci Monit 2018;24:1080-8. [Crossref] [PubMed]
- Liu Y, Li Y, Wang R, et al. MiR-130a-3p regulates cell migration and invasion via inhibition of SMAD4 in gemcitabine resistant hepatoma cells. J Exp Clin Cancer Res 2016;35:19. [Crossref] [PubMed]
- Cheng D, Zhao S, Tang H, et al. MicroRNA-20a-5p promotes colorectal cancer invasion and metastasis by downregulating SMAD4. Oncotarget 2016;7:45199-213. [Crossref] [PubMed]
- Wang Y, Ren J, Gao Y, et al. MicroRNA-224 targets SMAD family member 4 to promote cell proliferation and negatively influence patient survival. PLoS One 2013;8:e68744. [Crossref] [PubMed]
- Qiao P, Li G, Bi W, et al. microRNA-34a inhibits epithelial mesenchymal transition in human cholangiocarcinoma by targeting SMAD4 through transforming growth factor-beta/Smad pathway. BMC Cancer 2015;15:469. [Crossref] [PubMed]
- Jiang T, Ye L, Han Z, et al. miR-19b-3p promotes colon cancer proliferation and oxaliplatin-based chemoresistance by targeting SMAD4: validation by bioinformatics and experimental analyses. J Exp Clin Cancer Res 2017;36:131. [Crossref] [PubMed]
- Chai H, Liu M, Tian R, et al. miR-20a targets BNIP2 and contributes chemotherapeutic resistance in colorectal adenocarcinoma SW480 and SW620 cell lines. Acta Biochim Biophys Sin (Shanghai) 2011;43:217-25. [Crossref] [PubMed]
- Amankwatia EB, Chakravarty P, Carey FA, et al. MicroRNA-224 is associated with colorectal cancer progression and response to 5-fluorouracil-based chemotherapy by KRAS-dependent and -independent mechanisms. Br J Cancer 2015;112:1480-90. [Crossref] [PubMed]
- Kurokawa K, Tanahashi T, Iima T, et al. Role of miR-19b and its target mRNAs in 5-fluorouracil resistance in colon cancer cells. J Gastroenterol 2012;47:883-95. [Crossref] [PubMed]
- Sun C, Wang FJ, Zhang HG, et al. miR-34a mediates oxaliplatin resistance of colorectal cancer cells by inhibiting macroautophagy via transforming growth factor-b/SMAD4 pathway. World J Gastroenterol 2017;23:1816-27. [Crossref] [PubMed]
- Chung Y, Wi YC, Kim Y, et al. The SMAD4 /PTEN expression pattern predicts clinical outcomes in colorectal adenocarcinoma. J Pathol Transl Med 2018;52:37-44. [Crossref] [PubMed]
- Wei H, Cui R, Bahr J, et al. miR-130a deregulates PTEN and stimulates tumor growth. Cancer Res. 2017;77:6168-78. [Crossref] [PubMed]
- Wang D, Wang Y, Ma J, et al. MicroRNA-20a participates in the aerobic exercise-based prevention of coronary artery disease by targeting PTEN. Biomed Pharmacother 2017;95:756-63. [Crossref] [PubMed]
- Tian L, Fang YX, Xue JL, et al. Four microRNAs promote prostate cell proliferation with regulation of PTEN and its downstream signals in vitro. PLoS One 2013;8:e75885. [Crossref] [PubMed]
- Fleming NI, Jorissen RN, Mouradov D, et al. SMAD2, SMAD3 and SMAD4 mutations in colorectal cancer. Cancer Res 2013;73:725-35. [Crossref] [PubMed]
- Mehrvarz Sarshekeh A, Advani S, Overman MJ, et al. Association of SMAD4 mutation with patient demographics, tumor characteristics, and clinical outcomes in colorectal cancer. PLoS One 2017;12:e0173345. [Crossref] [PubMed]
- Yoshioka Y, Togashi Y, Chikugo T, et al. Clinicopathological and genetic differences between low-grade and high-grade colorectal mucinous adenocarcinomas. Cancer 2015;121:4359-68. [Crossref] [PubMed]
- Cerami E, Gao J, Dogrusoz U, et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov 2012;2:401-4. [Crossref] [PubMed]
- Dallol A, Buhmeida A, Al-Ahwal MS, et al. Clinical significance of frequent somatic mutations detected by high-throughput targeted sequencing in archived colorectal cancer samples. J Transl Med 2016;14:118. [Crossref] [PubMed]
- Zhang B, Zhang B, Chen X, et al. Loss of SMAD4 in colorectal cancer induces resistance to 5-fluorouracil through activating Akt pathway. Br J Cancer 2014;110:946-57. [Crossref] [PubMed]
- Mizuno T, Cloyd JM, Vicente D, et al. SMAD4 gene mutation predicts poor prognosis in patients undergoing resection for colorectal liver metastases. Eur J Surg Oncol 2018;44:684-92. [Crossref] [PubMed]
- Oba M, Hasegawa K, Matsuyama Y, et al. Discrepancy between recurrence-free survival and overall survival in patients with resectable colorectal liver metastases: a potential surrogate endpoint for time to surgical failure. Ann Surg Oncol 2014;21:1817-24. [Crossref] [PubMed]
- Oba M, Hasegawa K, Shindoh J, et al. Survival benefit of repeat resection of successive recurrences after the initial hepatic resection for colorectal liver metastases. Surgery 2016;159:632-40. [Crossref] [PubMed]
- Crespo P, Xu N, Simonds WF, et al. Ras-dependent activation of MAP kinase pathway mediated by G-protein beta gamma subunits. Nature 1994;369:418-20. [Crossref] [PubMed]
- Yamashita S, Chun YS, Kopetz SE, et al. APC and PIK3CA mutational cooperativity predicts pathologic response and survival in patients undergoing resection for colorectal liver metastases. Ann Surg 2017. [Epub ahead of print]. [Crossref] [PubMed]
- Løes IM, Immervoll H, Sorbye H, et al. Impact of KRAS, BRAF, PIK3CA, TP53 status and intraindividual mutation heterogeneity on outcome after liver resection for colorectal cancer metastases. Int J Cancer 2016;139:647-56. [Crossref] [PubMed]
- Kawaguchi Y, Kopetz S, Newhook TE, et al. Mutation Status of RAS, TP53, and SMAD4 is Superior to Mutation Status of RAS Alone for Predicting Prognosis after Resection of Colorectal Liver Metastases. Clin Cancer Res 2019. [Epub ahead of print]. [Crossref] [PubMed]


