Clinical utility of circulating Epstein-Barr virus DNA analysis for the management of nasopharyngeal carcinoma
Introduction
Nasopharyngeal carcinoma (NPC) is prevalent in Southern China and Southeast Asia. The annual incidence rate of NPC is 20–30 per 100,000 persons in Southern China compared with the world average of less than 1 per 100,000 persons (1). The incidence peaks at the age of 50 years and is two to three times more common in male than in females (2). Epstein-Barr virus (EBV) infection plays an important role in the pathogenesis of NPC (3). EBV genome can be detected in virtually all WHO Type III non-keratinizing, undifferentiated NPC tumors, the predominant subtype, in endemic areas (4).
Because of the close relationship between EBV and NPC, the possibility of using circulating EBV DNA as a tumor marker for NPC has been explored. Using PCR amplification followed by gel electrophoresis, Mutirangura et al. demonstrated that EBV DNA could be detected in the serum of 13 (31%) of 42 NPC patients but none of the 82 healthy subjects (5). This low detection rate in healthy subjects is remarkable given that over 95% of healthy adults have subclinical infections and EBV genome would reside in latently infected B lymphocytes (6). This finding suggests that the cell-associated EBV genome would not lead to false-positive results when cell-free plasma is analyzed (7). However, this method was not directly translated into clinical application because of the low sensitivity in detecting NPC patients. Circulating EBV DNA analysis has become clinically useful when Lo et al. demonstrated that EBV DNA could be detected in the plasma of 96% of NPC patients using real-time PCR analysis with a false-positive rate of about 7% (8). In this review, the clinical utilities of plasma EBV DNA for the detection, monitoring and prognostication of NPC will be discussed (Figure 1).
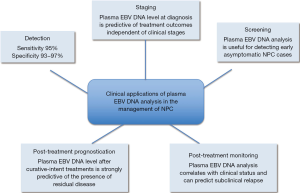
Detection of circulating Epstein-Barr virus (EBV) DNA in nasopharyngeal carcinoma (NPC) patients
With the high sensitivity and specificity of plasma EBV DNA analysis for NPC demonstrated by Lo et al. (8), this method have been further validated by different groups around the world. These validation studies varied in assay design, choice of sample type (serum vs. plasma), extraction volume and criteria for study subjects. The results of the representative studies are summarized in Table 1. The reported sensitivities ranged from 53% to 96% and the specificities ranged from 88% to 100%. Among four of the six studies reporting sensitivities of less than 90%, DNA was extracted from only 200 µL of serum/plasma (14-19). This relatively small extraction volume would limit the amount of extracted EBV DNA used for PCR analysis and adversely affect the detection rate. In addition, all the six studies showing low sensitivities involved assays targeting a single copy gene of the EBV genome (14-19). Those studies with reported sensitivities of greater than 90% involved assays targeting the BamHI-W fragments which has approximately ten repeats in an EBV genome (8,10-13). These results suggest that assays targeting a repeat region may potentially yield higher sensitivities. However, the two studies directly comparing the use of two assays each targeting a single copy gene and the BamHI-W fragment on the same patient cohort failed to confirm the superiority of the BamHI-W assay (9,16). This is likely due to the relatively small number of patients in those studies with only 19 and 23 patients were analyzed for the comparison (9,16). Moreover, only a few patients in the two cohorts were of early stage disease. The difference in sensitivities between these two types of assays should be more prominent when NPC patients with small tumor and low level of EBV DNA in plasma are studied.
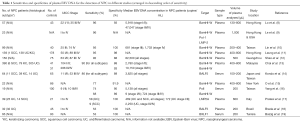
Full table
Another important factor that affects the detection rate is the clinical stage at diagnosis. Leung et al. showed that the detection rate of plasma EBV DNA was 90% and 98% for patients with early and late stage NPC respectively (11). The corresponding detection rates of viral capsid antigen (VCA) IgA were 72% and 85%. The superior sensitivity of EBV DNA over EBV IgA serology for the detection of NPC was later confirmed by other studies (14,20).
The histological subtypes of tumor is an another factor that would affect the detection rate of plasma EBV DNA in NPC patients. In endemic areas, most of the NPC are non-keratinizing and undifferentiated (WHO type III) and these tumors typically harbor the EBV genome (4). On the other hand, a significant proportion of type I NPCs (keratinizing squamous cell carcinoma) in non-endemic regions do not harbor EBV (21). In this regard, Pratesi et al. showed that the detection rate of plasma EBV DNA was lower in patients with squamous NPC compared to those with undifferentiated NPC (17).
Prognostication
In the first quantitative plasma EBV DNA study with 57 NPC patients by Lo et al. (8), the median concentrations of plasma EBV DNA was shown to be eight folds higher in patients with advanced stage (stages III and IV) compared with those with early stage disease (stages I and II) (8). This finding suggested that concentration of plasma EBV DNA would be able to reflect the tumor load in NPC patients. This hypothesis was confirmed by Ma et al. who demonstrated a positive relationship between plasma EBV DNA concentration at diagnosis and NPC tumor volume on magnetic resonance imaging in 57 patients with advanced stage NPC (22). The mathematical relationship between NPC tumor load and plasma EBV DNA level was further established using a mouse model (23). Chan et al. demonstrated a linear relationship between plasma EBV DNA concentration and tumor mass in nude mice engrafted with human NPC xenografts (23).
Because of the direct relationship between tumor load and plasma EBV DNA concentration, it is logical to deduce that plasma EBV DNA would be useful to predict the treatment outcomes of NPC patients. Lo et al. demonstrated that patients who developed local recurrence or distant metastasis within 1 year of curative-intent treatment had significantly higher pretreatment plasma EBV DNA concentrations than the group without recurrence or metastasis. Furthermore, they showed that the risk of disease-related mortality would be 1.6 folds for each 10-fold increase in pretreatment plasma EBV DNA concentration (24). Subsequent studies further showed the more powerful prognostic value of plasma EBV DNA in predicting overall survival than clinical staging of patients according to the Union for International Cancer Control (UICC) staging system (10,25,26). Leung et al. demonstrated that for NPC patients with stage I/II disease, those with high plasma EBV DNA at diagnosis had overall survival similar to stage III patients, whereas those with low plasma EBV DNA had overall survival approaching to that of stage I patients (10). In addition, Lin et al. showed that patients with high plasma EBV DNA were more likely to develop distant metastasis rather than locoregional recurrence (10). These results suggested that plasma EBV DNA analysis could be usefully incorporated into the staging system of NPC.
However, one practical challenge for incorporating EBV DNA into the staging system is the lack of standardization of assays for the measurement of the plasma EBV DNA concentration. While difference in assay design and extraction protocol would lead to difference in sensitivity and quantitative readouts, Le et al. further showed that solely adopting the same assay design could not eliminate the difference in the performance of the same assay in different laboratories. They demonstrated that the performance of different laboratories could only be harmonized with the standardization of all procedures involving sample collection, processing and result reporting, as well as the use of calibrators and qualitative assurance materials prepared from a single laboratory (27).
Detection of residual disease after treatment
Following treatment, cancer-derived EBV DNA would be rapidly cleared from the plasma of NPC patients. To et al. analyzed 21 patients with persistent or locoregional recurrent NPC who were treated with surgical resection of the tumor (28). They demonstrated that plasma EBV DNA was rapidly cleared from the circulation with a median half-life of only 139 minutes following the resection of the tumor (28). Renal excretion has been proposed to be one possible mechanism for the clearance as short EBV DNA fragments can be detected in the urine of NPC patients (29). The rapid clearance kinetics suggests that a huge amount of EBV DNA would be released from the tumor cells so as to maintain a high equilibrium level of plasma EBV DNA. For example, to maintain a plasma EBV DNA level of 5,000 copies/mL, which is the median level for patients with early NPC, over 100 million copies of EBV DNA would need to be released into the circulation every day (30). This raised the question on whether active viral replication is involved in the generation of this large amount of EBV DNA. In this regard, Chan et al. demonstrated that the EBV DNA in the circulation were mostly short fragments of less than 200 bp and were degradable with DNase digestion (31). These results suggest that the tumor-derived EBV DNA is only naked DNA fragments instead of associating with viral particles (31).
In addition to demonstrating the rapid clearance of plasma EBV DNA after NPC tumor resection, the clearance rate of EBV DNA from the plasma of NPC patients following radiotherapy was also studied (32). In patients receiving radiotherapy, the changes in plasma EBV DNA level would reflect the reduced production of EBV DNA from the tumor cells. Interestingly, a surge of plasma EBV DNA concentration was observed in every NPC patient immediately following the radiotherapy treatment (32). This rapid surge is due to the rapid release of EBV DNA from the dying NPC cells. After the surge, plasma EBV DNA concentration would reduce with a median half-life of 3.8 days (32). The rate of decline in plasma EBV DNA was later shown to be predictive of treatment outcomes. Patients with more rapid reduction were shown to have better survival than those with slower decline in plasma EBV DNA level (33-35). This observation suggests that the decline rate of plasma EBV DNA may reflect the radio-sensitivity of the tumor cells.
As EBV DNA is directly derived from the NPC tumor cells, the successful elimination of all tumor cells by curative treatment is expected to lead to the complete clearance of circulating EBV DNA. On the other hand, the persistence of EBV DNA in plasma after curative-intent treatment would suggest the presence of residual tumor cells in the body. In this regard, Chan et al. showed that the level of post-treatment EBV DNA was strongly predictive of overall and disease-free survival. Its predictive effect dominates the staging of the patients and the pretreatment plasma EBV DNA level (36). The relative risk for NPC recurrence was 11.9 for patients with post-treatment EBV DNA >500 copies/mL compared with those with undetectable level (36). These results were confirmed by studies with patient cohorts with different ethnic origins and clinical stages (10,37,38). Leung et al. further demonstrated that detectable plasma EBV DNA level at midpoint of the course of radiotherapy/chemoradiotherapy was associated with subsequent development of distant metastasis, worse overall and disease free survival compared with patients with undetectable level (35). Early identification of patients with high risk of disease recurrence by plasma EBV DNA analysis opens up the possibility of salvaging these patients with intensified chemotherapy whereas patients with undetectable plasma EBV DNA could be spared from further treatment to avoid treatment-associated side effects.
Screening for early disease
The most important parameter affecting the treatment outcome of NPC patients is the disease stage at diagnosis. The 5-year survival rate for patients with localized disease is up to 90% and the figure drops to below 50% for patients with metastatic disease (39). Unfortunately, more than three-quarters of NPC patients present late and have developed locally advanced or metastatic disease at presentation (25,39). Therefore, screening for early asymptomatic NPC may potentially improve the overall treatment outcome of these patients. In a proof-of-principle study, Chan et al. screened over 1,300 asymptomatic individuals of age 40–60 years for NPC using plasma EBV DNA analysis and EBV IgA serology (40). Subjects with positive results for either test were further investigated with nasal endoscopy examination. Three patients with early stage NPC (1 stage I and 2 stage II) were detected. All of them were positive for plasma EBV DNA but only one was positive for the EBV VCA IgA. These results suggest that plasma EBV DNA was more sensitive than EBV IgA serology in the NPC screening context (40). Interestingly, approximately 0.2% of subjects in the cohort had persistently positive plasma EBV DNA results but negative for NPC on endoscopy. A long-term follow up of these patients would be useful to clarify the clinical significance of persistently positive plasma EBV DNA results. In a previous study, subjects with positive EBV IgA serology were found to have a relative risk of 4 to 10 folds for developing NPC (41). It is possible that patients with persistently positive plasma EBV DNA may have increased risk of NPC development in the future. In addition to NPC, EBV DNA can be detected in other EBV-associated conditions, include infectious mononucleosis (42), Hodgkin disease (43) and Natural Killer/T-cell lymphoma (44) and post-transplant lymphoproliferative disorder (45). Therefore, these conditions need to be considered when plasma EBV DNA analysis is used as a tool for NPC screening.
Conclusions
Studies of plasma EBV DNA in NPC patients have highlighted the potential applications of the analysis cancer-derived DNA in the management of cancer patients, ranging from early cancer screening to the detection of residual disease after treatment. Over the last few years, other cancer-associated aberrations, including copy number alterations (46,47), single nucleotide mutations (48,49), translocations (50), methylation changes (51) and alteration of the size of circulating DNA (52,53), have been detected in patients with different types of cancers. It is expected that these new molecular markers will become important tools for patient management as in the model of EBV DNA in the management of NPC patients.
Acknowledgements
Funding: This work is supported by the Hong Kong Research Grants Council Theme-Based Research Scheme (T12-404/11) and Technology Fund under the State Key Laboratory Programme.
Footnote
Conflicts of Interest: KC Chan has patents/patent applications on molecular diagnostic methods. KC Chan is a director of Xcelom and Cirina. KC Chan has equity interests in Xcelom, Cirina and Sequenom. KC Chan is a consultant of Xcelom. The other authors have no conflicts of interest to declare.
References
- Wei WI, Sham JS. Nasopharyngeal carcinoma. Lancet 2005;365:2041-54. [Crossref] [PubMed]
- Registry HAHKC. Hong Kong Cancer Statistics Hong Kong: Hospital Authority, 2012 [cited 2015 20 October]. Available online: www3.ha.org.hk/cancereg/statistics.html
- Henle W, Henle G. The Epstein-Barr Virus (EBV) in Burkitt's lymphoma and nasopharyngeal carcinoma. Ann Clin Lab Sci 1974;4:109-14. [PubMed]
- Chen CL, Wen WN, Chen JY, et al. Detection of Epstein-Barr virus genome in nasopharyngeal carcinoma by in situ DNA hybridization. Intervirology 1993;36:91-8. [PubMed]
- Mutirangura A, Pornthanakasem W, Theamboonlers A, et al. Epstein-Barr viral DNA in serum of patients with nasopharyngeal carcinoma. Clin Cancer Res 1998;4:665-9. [PubMed]
- Hatton OL, Harris-Arnold A, Schaffert S, et al. The interplay between Epstein-Barr virus and B lymphocytes: implications for infection, immunity, and disease. Immunol Res 2014;58:268-76. [Crossref] [PubMed]
- Lit LC, Chan KC, Leung SF, et al. Distribution of cell-free and cell-associated Epstein-Barr virus (EBV) DNA in the blood of patients with nasopharyngeal carcinoma and EBV-associated lymphoma. Clin Chem 2004;50:1842-5. [Crossref] [PubMed]
- Lo YM, Chan LY, Lo KW, et al. Quantitative analysis of cell-free Epstein-Barr virus DNA in plasma of patients with nasopharyngeal carcinoma. Cancer Res 1999;59:1188-91. [PubMed]
- Le QT, Jones CD, Yau TK, et al. A comparison study of different PCR assays in measuring circulating plasma epstein-barr virus DNA levels in patients with nasopharyngeal carcinoma. Clin Cancer Res 2005;11:5700-7. [Crossref] [PubMed]
- Lin JC, Wang WY, Chen KY, et al. Quantification of plasma Epstein-Barr virus DNA in patients with advanced nasopharyngeal carcinoma. N Engl J Med 2004;350:2461-70. [Crossref] [PubMed]
- Leung SF, Tam JS, Chan AT, et al. Improved accuracy of detection of nasopharyngeal carcinoma by combined application of circulating Epstein-Barr virus DNA and anti-Epstein-Barr viral capsid antigen IgA antibody. Clin Chem 2004;50:339-45. [Crossref] [PubMed]
- Shao JY, Zhang Y, Li YH, et al. Comparison of Epstein-Barr virus DNA level in plasma, peripheral blood cell and tumor tissue in nasopharyngeal carcinoma. Anticancer Res 2004;24:4059-66. [PubMed]
- Chai SJ, Pua KC, Saleh A, et al. Clinical significance of plasma Epstein-Barr Virus DNA loads in a large cohort of Malaysian patients with nasopharyngeal carcinoma. J Clin Virol 2012;55:34-9. [Crossref] [PubMed]
- Kondo S, Horikawa T, Takeshita H, et al. Diagnostic value of serum EBV-DNA quantification and antibody to viral capsid antigen in nasopharyngeal carcinoma patients. Cancer Sci 2004;95:508-13. [Crossref] [PubMed]
- O TM, Yu G, Hu K, et al. Plasma Epstein-Barr virus immunoglobulin A and DNA for nasopharyngeal carcinoma screening in the United States. Otolaryngol Head Neck Surg 2007;136:992-7. [Crossref] [PubMed]
- Yang X, Goldstein AM, Chen CJ, et al. Distribution of Epstein-Barr viral load in serum of individuals from nasopharyngeal carcinoma high-risk families in Taiwan. Int J Cancer 2006;118:780-4. [Crossref] [PubMed]
- Pratesi C, Bortolin MT, D’Andrea M, et al. Quantitative plasma/serum EBV DNA load by LMP2A determination in an Italian cohort of NPC patients. J Clin Virol 2003;28:155-64. [Crossref] [PubMed]
- Breda E, Catarino RJ, Azevedo I, et al. Epstein-Barr virus detection in nasopharyngeal carcinoma: implications in a low-risk area. Braz J Otorhinolaryngol 2010;76:310-5. [Crossref] [PubMed]
- Baizig NM, Morand P, Seigneurin JM, et al. Complementary determination of Epstein-Barr virus DNA load and serum markers for nasopharyngeal carcinoma screening and early detection in individuals at risk in Tunisia. Eur Arch Otorhinolaryngol 2012;269:1005-11. [Crossref] [PubMed]
- Shao JY, Li YH, Gao HY, et al. Comparison of plasma Epstein-Barr virus (EBV) DNA levels and serum EBV immunoglobulin A/virus capsid antigen antibody titers in patients with nasopharyngeal carcinoma. Cancer 2004;100:1162-70. [Crossref] [PubMed]
- Nicholls JM, Agathanggelou A, Fung K, et al. The association of squamous cell carcinomas of the nasopharynx with Epstein-Barr virus shows geographical variation reminiscent of Burkitt's lymphoma. J Pathol 1997;183:164-8. [Crossref] [PubMed]
- Ma BB, King A, Lo YM, et al. Relationship between pretreatment level of plasma Epstein-Barr virus DNA, tumor burden, and metabolic activity in advanced nasopharyngeal carcinoma. Int J Radiat Oncol Biol Phys 2006;66:714-20. [Crossref] [PubMed]
- Chan KC, Chan AT, Leung SF, et al. Investigation into the origin and tumoral mass correlation of plasma Epstein-Barr virus DNA in nasopharyngeal carcinoma. Clin Chem 2005;51:2192-5. [Crossref] [PubMed]
- Lo YM, Chan AT, Chan LY, et al. Molecular prognostication of nasopharyngeal carcinoma by quantitative analysis of circulating Epstein-Barr virus DNA. Cancer Res 2000;60:6878-81. [PubMed]
- Leung SF, Zee B, Ma BB, et al. Plasma Epstein-Barr viral deoxyribonucleic acid quantitation complements tumor-node-metastasis staging prognostication in nasopharyngeal carcinoma. J Clin Oncol 2006;24:5414-8. [Crossref] [PubMed]
- Shen T, Tang LQ, Luo DH, et al. Different prognostic values of plasma Epstein-Barr virus DNA and maximal standardized uptake value of 18F-FDG PET/CT for nasopharyngeal carcinoma patients with recurrence. PLoS One 2015;10:e0122756. [Crossref] [PubMed]
- Le QT, Zhang Q, Cao H, et al. An international collaboration to harmonize the quantitative plasma epstein-barr virus DNA assay for future biomarker-guided trials in nasopharyngeal carcinoma. Clin Cancer Res 2013;19:2208-15. [Crossref] [PubMed]
- To EW, Chan KC, Leung SF, et al. Rapid clearance of plasma Epstein-Barr virus DNA after surgical treatment of nasopharyngeal carcinoma. Clin Cancer Res 2003;9:3254-9. [PubMed]
- Chan KC, Leung SF, Yeung SW, et al. Quantitative analysis of the transrenal excretion of circulating EBV DNA in nasopharyngeal carcinoma patients. Clin Cancer Res 2008;14:4809-13. [Crossref] [PubMed]
- Chan KC. Plasma Epstein-Barr virus DNA as a biomarker for nasopharyngeal carcinoma. Chin J Cancer 2014;33:598-603. [PubMed]
- Chan KC, Zhang J, Chan AT, et al. Molecular characterization of circulating EBV DNA in the plasma of nasopharyngeal carcinoma and lymphoma patients. Cancer Res 2003;63:2028-32. [PubMed]
- Lo YM, Leung SF, Chan LY, et al. Kinetics of plasma Epstein-Barr virus DNA during radiation therapy for nasopharyngeal carcinoma. Cancer Res 2000;60:2351-5. [PubMed]
- Hsu CL, Chang KP, Lin CY, et al. Plasma Epstein-Barr virus DNA concentration and clearance rate as novel prognostic factors for metastatic nasopharyngeal carcinoma. Head Neck 2012;34:1064-70. [Crossref] [PubMed]
- Wang WY, Twu CW, Chen HH, et al. Plasma EBV DNA clearance rate as a novel prognostic marker for metastatic/recurrent nasopharyngeal carcinoma. Clin Cancer Res 2010;16:1016-24. [Crossref] [PubMed]
- Leung SF, Chan KC, Ma BB, et al. Plasma Epstein-Barr viral DNA load at midpoint of radiotherapy course predicts outcome in advanced-stage nasopharyngeal carcinoma. Ann Oncol 2014;25:1204-8. [Crossref] [PubMed]
- Chan AT, Lo YM, Zee B, et al. Plasma Epstein-Barr virus DNA and residual disease after radiotherapy for undifferentiated nasopharyngeal carcinoma. J Natl Cancer Inst 2002;94:1614-9. [Crossref] [PubMed]
- An X, Wang FH, Ding PR, et al. Plasma Epstein-Barr virus DNA level strongly predicts survival in metastatic/recurrent nasopharyngeal carcinoma treated with palliative chemotherapy. Cancer 2011;117:3750-7. [Crossref] [PubMed]
- Ferrari D, Codeca C, Bertuzzi C, et al. Role of plasma EBV DNA levels in predicting recurrence of nasopharyngeal carcinoma in a Western population. BMC Cancer 2012;12:208. [Crossref] [PubMed]
- Lee AW, Sze WM, Au JS, et al. Treatment results for nasopharyngeal carcinoma in the modern era: the Hong Kong experience. Int J Radiat Oncol Biol Phys 2005;61:1107-16. [Crossref] [PubMed]
- Chan KC, Hung EC, Woo JK, et al. Early detection of nasopharyngeal carcinoma by plasma Epstein-Barr virus DNA analysis in a surveillance program. Cancer 2013;119:1838-44. [Crossref] [PubMed]
- Chien YC, Chen JY, Liu MY, et al. Serologic markers of Epstein-Barr virus infection and nasopharyngeal carcinoma in Taiwanese men. N Engl J Med 2001;345:1877-82. [Crossref] [PubMed]
- Pitetti RD, Laus S, Wadowsky RM. Clinical evaluation of a quantitative real time polymerase chain reaction assay for diagnosis of primary Epstein-Barr virus infection in children. Pediatr Infect Dis J 2003;22:736-9. [Crossref] [PubMed]
- Gandhi MK, Lambley E, Burrows J, et al. Plasma Epstein-Barr virus (EBV) DNA is a biomarker for EBV-positive Hodgkin's lymphoma. Clin Cancer Res 2006;12:460-4. [Crossref] [PubMed]
- Lei KI, Chan LY, Chan WY, et al. Quantitative analysis of circulating cell-free Epstein-Barr virus (EBV) DNA levels in patients with EBV-associated lymphoid malignancies. Br J Haematol 2000;111:239-46. [Crossref] [PubMed]
- Yang J, Tao Q, Flinn IW, et al. Characterization of Epstein-Barr virus-infected B cells in patients with posttransplantation lymphoproliferative disease: disappearance after rituximab therapy does not predict clinical response. Blood 2000;96:4055-63. [PubMed]
- Chan KC, Jiang P, Zheng YW, et al. Cancer genome scanning in plasma: Detection of tumor-associated copy number aberrations, single-nucleotide variants, and tumoral heterogeneity by massively parallel sequencing. Clin Chem 2013;59:211-24. [Crossref] [PubMed]
- Chan KC. Scanning for cancer genomic changes in plasma: toward an era of personalized blood-based tumor markers. Clin Chem 2013;59:1553-5. [Crossref] [PubMed]
- Yung TK, Chan KC, Mok TS, et al. Single-molecule detection of epidermal growth factor receptor mutations in plasma by microfluidics digital PCR in non-small cell lung cancer patients. Clin Cancer Res 2009;15:2076-84. [Crossref] [PubMed]
- Forshew T, Murtaza M, Parkinson C, et al. Noninvasive identification and monitoring of cancer mutations by targeted deep sequencing of plasma DNA. Sci Transl Med 2012;4:136ra68.
- Leary RJ, Kinde I, Diehl F, et al. Development of personalized tumor biomarkers using massively parallel sequencing. Sci Transl Med 2010;2:20ra14. [Crossref] [PubMed]
- Chan KC, Jiang P, Chan CW, et al. Noninvasive detection of cancer-associated genomewide hypomethylation and copy number aberrations by plasma DNA bisulfite sequencing. Proc Natl Acad Sci U S A 2013;110:18761-8. [Crossref] [PubMed]
- Chan KC, Leung SF, Yeung SW, et al. Persistent aberrations in circulating DNA integrity after radiotherapy are associated with poor prognosis in nasopharyngeal carcinoma patients. Clin Cancer Res 2008;14:4141-5. [Crossref] [PubMed]
- Jiang P, Chan CW, Chan KC, et al. Lengthening and shortening of plasma DNA in hepatocellular carcinoma patients. Proc Natl Acad Sci U S A 2015;112:E1317-25. [Crossref] [PubMed]


