
Interactions of multiple gene alterations in colorectal liver metastases
Introduction
Colorectal cancer is genetically heterogenous (1). Approximately 15–30% of patients with colorectal cancer develop colorectal liver metastases (CLM). Liver resection is regarded as a potentially curative treatment for patients with CLM and provides 5-year overall survival (OS) rates of approximately 40–60% (2-4). However, we experience clinical heterogeneity in patients undergoing CLM resection. Some patients have a good prognosis after CLM resection and achieve a prolonged cancer-free interval or a cure, while others have a progressive fatal disease. The recent development of the next generation sequencing has led to increased availability of gene alteration information in clinic practice. RAS alteration initially became the main focus because it determines resistance to anti-epidermal growth factor receptor (EGFR) agents (5-7). Furthermore, the prognostic role of alterations in RAS and BRAF has been well studied in patients with colorectal cancer (8-13). However, prognostic roles of gene alterations other than RAS and BRAF remain unclear. This article reviews the association of multiple gene alterations with prognosis after CLM resection to be useful for clinical decision-making.
Frequency of somatic gene alteration in patients with CLM
We recently reported 507 patients who had genetic sequencing data of 50 cancer-related genes and underwent CLM resection (14). Of the 50 genes analyzed, alterations of the following 13 genes were found in more than 1% of patients: TP53, APC, KRAS, PIK3CA, SMAD4, FBXW7, NRAS, ERBB2, ATM, BRAF, PTEN, RB1, and CTNNB1 (Table 1). The frequencies of gene alterations are similar to a project from the Cancer Genome Atlas Network (1). Alterations in TP53, APC, RAS (KRAS + NRAS) occur in more than 50% of patients with colorectal cancer. PIK3CA, SMDA4, and FBXW7 are the second most frequently altered gene group, ranging from 5% to 20% of alterations in this patient group.
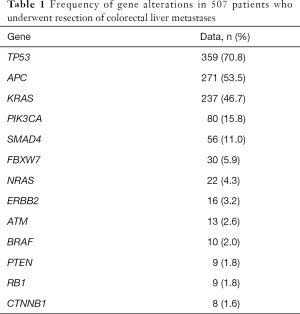
Full table
Alterations in RAS and BRAF and prognosis after CLM resection
Chemotherapy regimens, including anti-EGFR agents, have demonstrated improved survival in patients with unresectable metastatic colorectal cancer (15). Alterations in the RAS gene family (KRAS, NRAS, and HRAS) were found to be an important biomarker and used clinically to determine response to anti-EGFR therapy (5-7). The advancements in molecular-targeted therapy have quickly drawn attention to information on somatic alterations in colorectal cancer.
Studies have assessed an association of RAS alteration with prognosis in patients undergoing CLM resection (9,11,14,16-24). Most studies show worse OS and recurrence-free survival (RFS) in RAS altered patients than in RAS wild-type patients (9,11,14,23). In contrast, other studies have not shown significant differences between RAS altered patients and RAS wild-type patients in OS (17,18,21), or in RFS (20,21), using a multivariable Cox model.
Approximately 2–5% of patients undergoing CLM resection have BRAF alteration (12-14). Studies have reported that BRAF alteration occurs in approximately 10% of all patients with colorectal cancer (25). Moreover, BRAF altered patients with colorectal cancer were associated with worse prognosis (8,10). However, the rarity of BRAF altered patients undergoing CLM resection makes it hard to assess the prognostic role of BRAF alteration with sufficient statistical power. Recent multi-institutional studies evaluated 35 patients with BRAF alteration out of 1,497 total patients (12), and 45 patients with BRAF alteration out of 853 patients (13). Both studies demonstrated that OS and RFS were significantly worse in BRAF altered patients than in BRAF wild-type patients (12,13).
Alterations in TP53 and prognosis after CLM resection
TP53 is a tumor suppressor gene in the p53 pathway and has a role to inhibit tumor cell growth (26,27). The prognostic role of TP53 alteration in patients undergoing CLM resection has been previously studied in early 2000s (28-34). While four studies failed to show the association of TP53 alteration with prognosis (30,31,33,34), there are four other studies which showed that TP53 altered patients were associated with worse OS than TP53 wild-type patients (28,32,35). One study has shown better OS and RFS in TP53 altered patients than in TP53 wild-type patients (29). Because the type of gene alteration influences the change of proteins and functions, Chun et al. analyzed patients with TP53 altered patients (24) by classifying a risk of missense TP53 alteration into high and low risk groups on the basis of the evolutionary action score (36,37). TP53 alteration with a high evolutionary action score is associated with worse survival (24). OS and RFS were worse in patients with co-alteration in RAS and TP53 than in patients with one alteration of the two genes and in patients with no alteration (24,38).
Multiple alterations and prognosis after CLM resection
We recently analyzed prognostic roles of five frequently altered genes (TP53, APC, RAS, PIK3CA, SMAD4) as well as BRAF in 507 patients undergoing CLM resection (14). According to a multivariable Cox proportional hazards model analysis, alterations in RAS, TP53, and SMAD4 were significantly associated with worse OS and RFS, together with other clinicopathologic factors. BRAF alteration was an independent risk factor for OS but not for RFS. Since alterations in RAS, TP53, and SMAD4 were independently associated with OS and RFS, we calculated multivariable hazard ratios (HR) focusing on number of alterations among these three genes. Co-existing alterations of all three genes (triple alteration) was significantly associated with worse OS than co-existing alteration in any two of the genes (double alteration) [HR 3.21, 95% confidence interval (CI): 1.72–5.99, P<0.001]. Triple alteration was also significantly associated with worse OS than alteration in only one of the genes (single alteration) (HR 6.04, 95% CI: 3.21–11.3, P<0.001) and no alteration in any of the genes (all wild-type) (HR 8.61, 95% CI: 3.80–19.5, P<0.001). Similarly, triple alteration was significantly associated with worse RFS than double alteration (HR 2.06, 95% CI: 1.28–3.29, P=0.003), single alteration (HR 3.17, 95% CI: 1.97–5.07, P<0.001), and all wild-type (HR 3.72, 95% CI: 2.14–6.46, P<0.001). Furthermore, OS and RFS did not differ significantly between patients with single alteration vs. patients with all wild-type (OS, HR 1.43, 95% CI: 0.77–2.63, P=0.256; RFS, HR 1.18, 95% CI: 0.82–1.68, P=0.378). Based on this finding, we compared OS and RFS between patients with RAS alteration and wild-type TP53 and SMAD4 and patients with RAS wild-type. OS and RFS did not differ significantly between both patient groups: OS, HR 0.95, 95% CI: 0.55–1.65, P=0.858; RFS, HR 1.06, 95% CI: 0.77–1.44, P=0.729. Our findings likely explain why some studies found prognostic role of RAS alteration (9,11,14,23) while others failed to show an association of RAS alteration with prognosis (17,18,20,21). Overall, the testing of multiple gene alterations, including the analysis of co-existing alterations, may provide additional prognostic information in patients undergoing CLM resection.
RAS, TP53, and SMAD4 belong to different cancer-related signaling pathways (i.e., MAPK pathway, p53 pathway, and TGFβ pathway). Our recent study suggests that through the corresponding signaling pathways, the deleterious effects on survival are accumulated from single to double alterations, and also from double to triple alterations. However, our study cannot explain the interactions that may exist between these alterations. Studies report that there may exist an interaction and synergism for progression of carcinoma between MAPK pathway, p53 pathway, and TGFβ pathway. These interactions may in part account for the worse prognosis of patients with multiple gene alterations (39-41).
Conclusions
In patients with colorectal cancer, alterations in RAS and BRAF have been increasingly examined as biomarkers for testing resistance to anti-EGFR agents. Advancements in next generation sequencing have made multiple gene testing clinically available. Because RAS alteration status alone is not sufficient to predict survival after CLM resection, information on multiple gene alteration status may aid in predicting prognosis more precisely; and therefore, influence the clinical decision-making process. Further developments in other molecular targeted therapies may further highlight the importance of information on gene sequencing.
Acknowledgments
The authors thank Ms. Ruth Haynes for administrative support in the preparation of this manuscript.
Funding: This article was supported in part by the National Institutes of Health (T32 CA 009599) and the MD Anderson Cancer Center Support Grant (CA016672).
Footnote
Conflicts of Interest: The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
References
- Cancer Genome Atlas Network. Comprehensive molecular characterization of human colon and rectal cancer. Nature 2012;487:330-7. [Crossref] [PubMed]
- Choti MA, Sitzmann JV, Tiburi MF, et al. Trends in long-term survival following liver resection for hepatic colorectal metastases. Ann Surg 2002;235:759-66. [Crossref] [PubMed]
- Abdalla EK, Vauthey JN, Ellis LM, et al. Recurrence and Outcomes Following Hepatic Resection, Radiofrequency Ablation, and Combined Resection/Ablation for Colorectal Liver Metastases. Ann Surg 2004;239:818-27. [Crossref] [PubMed]
- Fernandez FG, Drebin JA, Linehan DC, et al. Five-Year Survival After Resection of Hepatic Metastases From Colorectal Cancer in Patients Screened by Positron Emission Tomography With F-18 Fluorodeoxyglucose (FDG-PET). Ann Surg 2004;240:438-50. [Crossref] [PubMed]
- Moroni M, Veronese S, Benvenuti S, et al. Gene copy number for epidermal growth factor receptor (EGFR) and clinical response to antiEGFR treatment in colorectal cancer: a cohort study. Lancet Oncol 2005;6:279-86. [Crossref] [PubMed]
- Van Cutsem E, Peeters M, Siena S, et al. Open-label phase III trial of panitumumab plus best supportive care compared with best supportive care alone in patients with chemotherapy-refractory metastatic colorectal cancer. J Clin Oncol 2007;25:1658-64. [Crossref] [PubMed]
- Amado RG, Wolf M, Peeters M, et al. Wild-type KRAS is required for panitumumab efficacy in patients with metastatic colorectal cancer. J Clin Oncol 2008;26:1626-34. [Crossref] [PubMed]
- Roth AD, Tejpar S, Delorenzi M, et al. Prognostic role of KRAS and BRAF in stage II and III resected colon cancer: results of the translational study on the PETACC-3, EORTC 40993, SAKK 60-00 trial. J Clin Oncol 2010;28:466-74. [Crossref] [PubMed]
- Vauthey JN, Zimmitti G, Kopetz SE, et al. RAS mutation status predicts survival and patterns of recurrence in patients undergoing hepatectomy for colorectal liver metastases. Ann Surg 2013;258:619-26; discussion 626-7. [Crossref] [PubMed]
- Yuan ZX, Wang XY, Qin QY, et al. The prognostic role of BRAF mutation in metastatic colorectal cancer receiving anti-EGFR monoclonal antibodies: a meta-analysis. PLoS One 2013;8:e65995. [Crossref] [PubMed]
- Karagkounis G, Torbenson MS, Daniel HD, et al. Incidence and prognostic impact of KRAS and BRAF mutation in patients undergoing liver surgery for colorectal metastases. Cancer 2013;119:4137-44. [Crossref] [PubMed]
- Gagniere J, Dupre A, Gholami SS, et al. Is Hepatectomy Justified for BRAF Mutant Colorectal Liver Metastases?: A Multi-institutional Analysis of 1497 Patients. Ann Surg 2018. [Epub ahead of print]. [Crossref] [PubMed]
- Margonis GA, Buettner S, Andreatos N, et al. Association of BRAF Mutations With Survival and Recurrence in Surgically Treated Patients With Metastatic Colorectal Liver Cancer. JAMA Surg 2018;153:e180996. [Crossref] [PubMed]
- Kawaguchi Y, Kopetz S, Newhook TE, et al. Mutation Status of RAS, TP53, and SMAD4 is Superior to Mutation Status of RAS Alone for Predicting Prognosis after Resection of Colorectal Liver Metastases. Clin Cancer Res 2019. [Epub ahead of print]. [Crossref] [PubMed]
- Cunningham D, Humblet Y, Siena S, et al. Cetuximab monotherapy and cetuximab plus irinotecan in irinotecan-refractory metastatic colorectal cancer. N Engl J Med 2004;351:337-45. [Crossref] [PubMed]
- Nash GM, Gimbel M, Shia J, et al. KRAS mutation correlates with accelerated metastatic progression in patients with colorectal liver metastases. Ann Surg Oncol 2010;17:572-8. [Crossref] [PubMed]
- Teng HW, Huang YC, Lin JK, et al. BRAF mutation is a prognostic biomarker for colorectal liver metastasectomy. J Surg Oncol 2012;106:123-9. [Crossref] [PubMed]
- Lin Q, Ye Q, Zhu D, et al. Determinants of long-term outcome in patients undergoing simultaneous resection of synchronous colorectal liver metastases. PLoS One 2014;9:e105747. [Crossref] [PubMed]
- Brudvik KW, Kopetz SE, Li L, et al. Meta-analysis of KRAS mutations and survival after resection of colorectal liver metastases. Br J Surg 2015;102:1175-83. [Crossref] [PubMed]
- Margonis GA, Spolverato G, Kim Y, et al. Effect of KRAS Mutation on Long-Term Outcomes of Patients Undergoing Hepatic Resection for Colorectal Liver Metastases. Ann Surg Oncol 2015;22:4158-65. [Crossref] [PubMed]
- Schirripa M, Bergamo F, Cremolini C, et al. BRAF and RAS mutations as prognostic factors in metastatic colorectal cancer patients undergoing liver resection. Br J Cancer 2015;112:1921-8. [Crossref] [PubMed]
- Wang K, Liu W, Yan XL, et al. Long-term postoperative survival prediction in patients with colorectal liver metastasis. Oncotarget 2017;8:79927-34. [PubMed]
- Amikura K, Akagi K, Ogura T, et al. The RAS mutation status predicts survival in patients undergoing hepatic resection for colorectal liver metastases: The results from a genetic analysis of all-RAS. J Surg Oncol 2018;117:745-55. [Crossref] [PubMed]
- Chun YS, Passot G, Yamashita S, et al. Deleterious Effect of RAS and Evolutionary High-risk TP53 Double Mutation in Colorectal Liver Metastases. Ann Surg 2019;269:917-23. [Crossref] [PubMed]
- Tejpar S, Bertagnolli M, Bosman F, et al. Prognostic and predictive biomarkers in resected colon cancer: current status and future perspectives for integrating genomics into biomarker discovery. Oncologist 2010;15:390-404. [Crossref] [PubMed]
- Vousden KH, Lu X. Live or let die: the cell's response to p53. Nat Rev Cancer 2002;2:594-604. [Crossref] [PubMed]
- Balint EE, Vousden KH. Activation and activities of the p53 tumour suppressor protein. Br J Cancer 2001;85:1813-23. [Crossref] [PubMed]
- Tullo A, D'Erchia AM, Honda K, et al. Characterization of p53 mutations in colorectal liver metastases and correlation with clinical parameters. Clin Cancer Res 1999;5:3523-8. [PubMed]
- Yang Y, Forslund A, Remotti H, et al. P53 mutations in primary tumors and subsequent liver metastases are related to survival in patients with colorectal carcinoma who undergo liver resection. Cancer 2001;91:727-36. [Crossref] [PubMed]
- Saw RP, Koorey D, Painter D, et al. p53, DCC and thymidylate synthase as predictors of survival after resection of hepatic metastases from colorectal cancer. Br J Surg 2002;89:1409-15. [Crossref] [PubMed]
- de Jong KP, Gouw AS, Peeters PM, et al. P53 mutation analysis of colorectal liver metastases: relation to actual survival, angiogenic status, and p53 overexpression. Clin Cancer Res 2005;11:4067-73. [Crossref] [PubMed]
- Pilat N, Grunberger T, Langle F, et al. Assessing the TP53 marker type in patients treated with or without neoadjuvant chemotherapy for resectable colorectal liver metastases: a p53 Research Group study. Eur J Surg Oncol 2015;41:683-9. [Crossref] [PubMed]
- Loes IM, Immervoll H, Sorbye H, et al. Impact of KRAS, BRAF, PIK3CA, TP53 status and intraindividual mutation heterogeneity on outcome after liver resection for colorectal cancer metastases. Int J Cancer 2016;139:647-56. [Crossref] [PubMed]
- Frankel TL, Vakiani E, Nathan H, et al. Mutation location on the RAS oncogene affects pathologic features and survival after resection of colorectal liver metastases. Cancer 2017;123:568-75. [Crossref] [PubMed]
- Mollevi DG, Serrano T, Ginesta MM, et al. Mutations in TP53 are a prognostic factor in colorectal hepatic metastases undergoing surgical resection. Carcinogenesis 2007;28:1241-6. [Crossref] [PubMed]
- Neskey DM, Osman AA, Ow TJ, et al. Evolutionary Action Score of TP53 Identifies High-Risk Mutations Associated with Decreased Survival and Increased Distant Metastases in Head and Neck Cancer. Cancer Res 2015;75:1527-36. [Crossref] [PubMed]
- Katsonis P, Lichtarge O. A formal perturbation equation between genotype and phenotype determines the Evolutionary Action of protein-coding variations on fitness. Genome Res 2014;24:2050-8. [Crossref] [PubMed]
- Kawaguchi Y, Lillemoe HA, Panettieri E, et al. Conditional Recurrence-Free Survival after Resection of Colorectal Liver Metastases: Persistent Deleterious Association with RAS and TP53 Co-Mutation. J Am Coll Surg 2019. [Epub ahead of print]. [Crossref] [PubMed]
- Grusch M, Petz M, Metzner T, et al. The crosstalk of RAS with the TGF-beta family during carcinoma progression and its implications for targeted cancer therapy. Curr Cancer Drug Targets 2010;10:849-57. [Crossref] [PubMed]
- Elston R, Inman GJ. Crosstalk between p53 and TGF-beta Signalling. J Signal Transduct 2012;2012:294097.
- Buganim Y, Solomon H, Rais Y, et al. p53 Regulates the Ras circuit to inhibit the expression of a cancer-related gene signature by various molecular pathways. Cancer Res 2010;70:2274-84. [Crossref] [PubMed]


